SynMap: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
|||
| Line 18: | Line 18: | ||
[[Image:SynPlot-Chr1-At-v-Al.png|thumb|right|600px|Syntenic dotplot and synonymous substitution histogram of Chromosome 1 of Arabidopsis thaliana and Arabidopsis lyrata. Syntenic gene pairs were identified by DAGChainer, and colored based on their synonymous substitution rate as calculated by CodeML. Syntenic regions derived from speciation (orthologs) or from the shared whole genome duplication events (alpha and beta) are labeled. These images were generated by SynMap and can be viewed at generated by SynMap at http://toxic.berkeley.edu/CoGe//diags/Arabidopsis_lyrata/Arabidopsis_thaliana_thale_cress/html/3068_8.CDS-CDS.blastn.dag_geneorder_D20_g10_A5.scaffold_1-1.w600.ks.html]] | [[Image:SynPlot-Chr1-At-v-Al.png|thumb|right|600px|Syntenic dotplot and synonymous substitution histogram of Chromosome 1 of Arabidopsis thaliana and Arabidopsis lyrata. Syntenic gene pairs were identified by DAGChainer, and colored based on their synonymous substitution rate as calculated by CodeML. Syntenic regions derived from speciation (orthologs) or from the shared whole genome duplication events (alpha and beta) are labeled. These images were generated by SynMap and can be viewed at generated by SynMap at http://toxic.berkeley.edu/CoGe//diags/Arabidopsis_lyrata/Arabidopsis_thaliana_thale_cress/html/3068_8.CDS-CDS.blastn.dag_geneorder_D20_g10_A5.scaffold_1-1.w600.ks.html]] | ||
This example shows a whole genome syntenic dotplot comparison of ''Arabidopsis thaliana'' (At) and ''Arabidopsis lyrata'' (Al). These taxa diverged from one another ~5MYA | This example shows a whole genome syntenic dotplot comparison of ''Arabidopsis thaliana'' (At) and ''Arabidopsis lyrata'' (Al). These taxa diverged from one another ~5MYA <ref name="lysak2006>Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proceedings of the National Academy of Sciences of the United States of America. 2006 Mar 28;103(13):5224-5229. | ||
</ref> | |||
<references/> | |||
{{reflist}} | |||
Revision as of 17:03, 28 August 2009
Overview
Specifying genomes
DAGChainer options
SynMap options
Calculating and displaying synonymous/non-synonymous (kS, kN) data
Interacting with results
Example Results

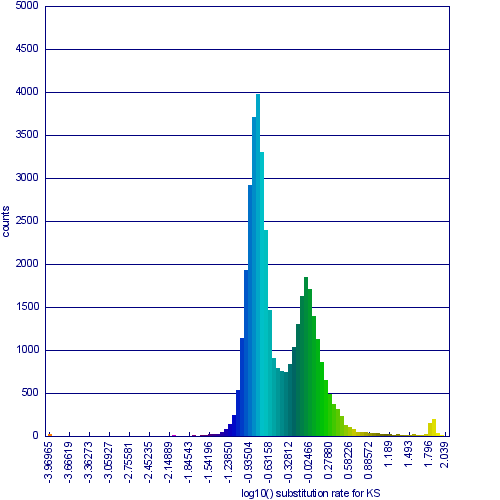
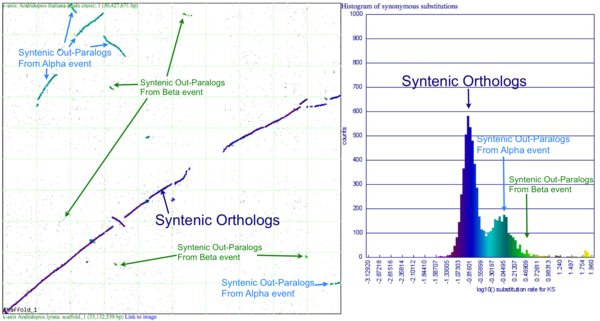
This example shows a whole genome syntenic dotplot comparison of Arabidopsis thaliana (At) and Arabidopsis lyrata (Al). These taxa diverged from one another ~5MYA [1]
- ↑ Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proceedings of the National Academy of Sciences of the United States of America. 2006 Mar 28;103(13):5224-5229.