Ancestral Reconstruction Pipeline: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 12: | Line 12: | ||
* -s < subgenome_file> | * -s < subgenome_file> | ||
* -o <output directory for SubGenomeInGeneOrder OrthologSets GenomeInString files> | * -o <output directory for SubGenomeInGeneOrder OrthologSets GenomeInString files> | ||
====GetContigInput==== | ====GetContigInput==== | ||
*Remove config file dependency | *Remove config file dependency | ||
| Line 25: | Line 20: | ||
*-b <Number of bins for assignment to ancestral chromosomes. Called 'AncChrNumber' in original config file> (Note: this may be removed if this info can be derived from the input.> | *-b <Number of bins for assignment to ancestral chromosomes. Called 'AncChrNumber' in original config file> (Note: this may be removed if this info can be derived from the input.> | ||
Note: genomeInContigIndex is specified in the config file. This is the weighting for each subgenome. Need to discuss how best to deal with these. | Note: genomeInContigIndex is specified in the config file. This is the weighting for each subgenome. Need to discuss how best to deal with these. | ||
====MWMPython: http://jorisvr.nl/maximummatching.html needs command line options for ==== | |||
* -i <input file or directory> | |||
*** File type is a set of vertex vertex weight | |||
*** note: if directory, will batch process all files | |||
* -o <outfile or directory> | |||
Revision as of 21:36, 25 April 2014
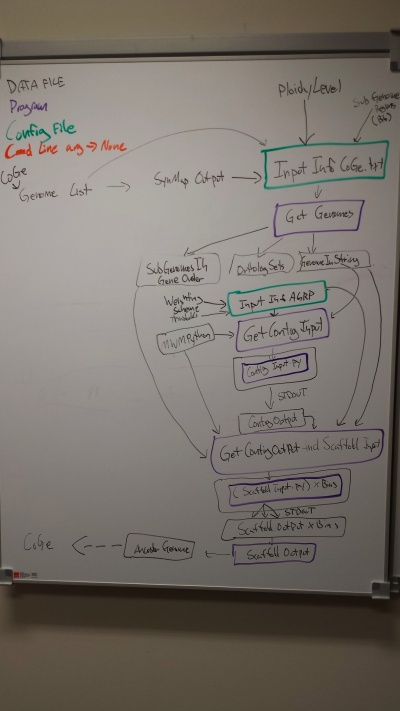
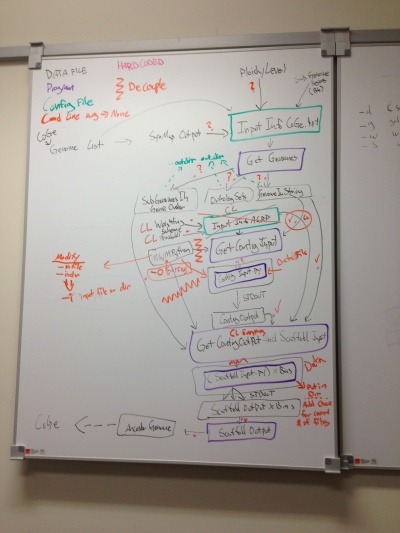
Plan for refactoring
GetGenomes: remove config file, add option to specify output dir for output files
- -d<directory of input synmap files>
- -g gid1,gid2,gid3,gid4... <list of common separated coge genome ids>
- -w w1,w2,w3,w4 <list of comma separated syntenic depths for genomes -- note these are paired ordered data with the -g option?>
- -s < subgenome_file>
- -o <output directory for SubGenomeInGeneOrder OrthologSets GenomeInString files>
GetContigInput
- Remove config file dependency
- -g gid1,gid2,gid3,gid4... <list of common separated coge genome ids>
- -w w1,w2,w3,w4 <list of comma separated weights for genomes -- note these are paired ordered data with the -g option?>
- -wa <threshold minimum adjacency score for keeping a contig. Called 'weightOfAdjacent' in original config file>
- -cl <threshold minimum contig length. Called 'minimumGeneGroupLength' in original config file>
- -b <Number of bins for assignment to ancestral chromosomes. Called 'AncChrNumber' in original config file> (Note: this may be removed if this info can be derived from the input.>
Note: genomeInContigIndex is specified in the config file. This is the weighting for each subgenome. Need to discuss how best to deal with these.
MWMPython: http://jorisvr.nl/maximummatching.html needs command line options for
- -i <input file or directory>
- File type is a set of vertex vertex weight
- note: if directory, will batch process all files
- -o <outfile or directory>