Expression Analysis Pipeline: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 5: | Line 5: | ||
When a FASTQ file of sequence reads is loaded in [[LoadExperiment]] and associated with an annotated genome, the following analysis steps are performed: | When a FASTQ file of sequence reads is loaded in [[LoadExperiment]] and associated with an annotated genome, the following analysis steps are performed: | ||
# The FASTQ file is verified for correct format. | # The FASTQ file is verified for correct format. | ||
# [http://code.google.com/p/cutadapt/ CutAdapt] is run to trim adapter sequence from the reads (parameters: -q 25 | # [http://code.google.com/p/cutadapt/ CutAdapt] is run to trim adapter sequence from the reads (parameters: -q 25 -m 17). | ||
# [http://research-pub.gene.com/gmap/ GMAP] or [http://bowtie-bio.sourceforge.net/bowtie2/index.shtml Bowtie2] is run to index the reference genome sequence, depending on your choice. | # [http://research-pub.gene.com/gmap/ GMAP] or [http://bowtie-bio.sourceforge.net/bowtie2/index.shtml Bowtie2] is run to index the reference genome sequence, depending on your choice. | ||
# [http://research-pub.gene.com/gmap/ GSNAP] or [http://tophat.cbcb.umd.edu/ TopHat] is run to align the reads to the reference sequence (GSNAP parameters: -n 5 --format=sam -Q --gmap-mode=none --nofails, TopHat parameters: -g 1). | # [http://research-pub.gene.com/gmap/ GSNAP] or [http://tophat.cbcb.umd.edu/ TopHat] is run to align the reads to the reference sequence (GSNAP parameters: -n 5 --format=sam -Q --gmap-mode=none --nofails, TopHat parameters: -g 1). | ||
| Line 16: | Line 16: | ||
TBD: how to do this ... | TBD: how to do this ... | ||
Note: | |||
* Removed "--quality-base=64" as an argument for cutadapt from the original qteller pipeline | |||
===Video Tutorial=== | ===Video Tutorial=== | ||
Revision as of 17:26, 7 May 2014

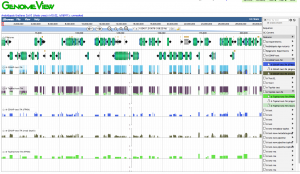
CoGe can generate gene/transcript expression measurements given a FASTQ input and an annotated genome. Thanks to James Schnable, creator of qTeller, for help developing this pipeline!
When a FASTQ file of sequence reads is loaded in LoadExperiment and associated with an annotated genome, the following analysis steps are performed:
- The FASTQ file is verified for correct format.
- CutAdapt is run to trim adapter sequence from the reads (parameters: -q 25 -m 17).
- GMAP or Bowtie2 is run to index the reference genome sequence, depending on your choice.
- GSNAP or TopHat is run to align the reads to the reference sequence (GSNAP parameters: -n 5 --format=sam -Q --gmap-mode=none --nofails, TopHat parameters: -g 1).
- SAMtools is run to compute per-position read depth of the resulting alignment (mpileup -D -Q 20).
- Cufflinks is run to compte per-transcript FPKM (parameters: -p 24).
- The per-position read depth and per-transcript FPKM values are log transformed and normalized between 0 and 1 for loading.
- The three results (raw alignment, per-position read depth, and per-transcript FPKM) are loaded as separate Experiments into a Notebook.
Genomes for which this analysis has been performed can have features imported into qTeller. TBD: how to do this ...
Note:
- Removed "--quality-base=64" as an argument for cutadapt from the original qteller pipeline
Video Tutorial
- Demo fastq data for Arabidopsis Col-0:
- 0.17M reads: http://de.iplantcollaborative.org/dl/d/2F807292-34CC-4C8E-96E3-3E668A304D23/test_rna_seq_data_0.17M_reads.fastq
- 1M reads: http://de.iplantcollaborative.org/dl/d/EFD4F983-80B1-4388-94C4-AD78E73D2795/test_rna_seq_data_1M_reads.fastq
- 7.6M reads: http://de.iplantcollaborative.org/dl/d/9F6602D6-C66B-4C97-A72A-180AAE55AF95/test_rna_seq_data_7.6M_reads.fastq