GFF ingestion: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 4: | Line 4: | ||
*[[CDS]]: the regions that code for protein. | *[[CDS]]: the regions that code for protein. | ||
[[File:Screen shot 2012-04-17 at 1.10.42 PM.png | [[File:Screen shot 2012-04-17 at 1.10.42 PM.png|right|400px|CoGe visualization of [[genomic feature]] from the rice genome]] | ||
From the GFF3 entry below, the gene and mRNA features are collapsed to a gene in CoGe, the exons are combined to make an mRNA in CoGe, and the CDSs are used as a CDS feature in CoGe. The UTRs are skipped as being redundant with the exons. | From the GFF3 entry below, the gene and mRNA features are collapsed to a gene in CoGe, the exons are combined to make an mRNA in CoGe, and the CDSs are used as a CDS feature in CoGe. The UTRs are skipped as being redundant with the exons. | ||
Revision as of 23:48, 7 July 2014
CoGe translates many of the features from a standard GFF file into various genomic features in CoGe's database. For a basic protein coding gene, CoGe tracks three major genomic features:
- gene: the full extent of the transcribed unit including introns
- mRNA: the spliced transcript
- CDS: the regions that code for protein.
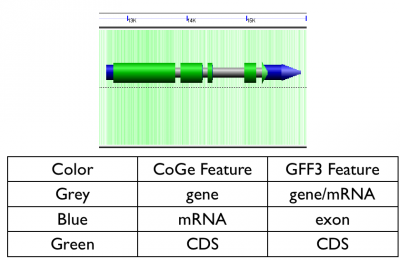
From the GFF3 entry below, the gene and mRNA features are collapsed to a gene in CoGe, the exons are combined to make an mRNA in CoGe, and the CDSs are used as a CDS feature in CoGe. The UTRs are skipped as being redundant with the exons.
Example GFF entry for a protein coding gene from the rice genome (v7)
Chr1 MSU_osa1r7 gene 12648 15915 . + . ID=LOC_Os01g01030;Name=LOC_Os01g01030;Note=monocopper%20oxidase%2C%20putative%2C%20expressed Chr1 MSU_osa1r7 mRNA 12648 15915 . + . ID=LOC_Os01g01030.1;Name=LOC_Os01g01030.1;Parent=LOC_Os01g01030 Chr1 MSU_osa1r7 exon 12648 13813 . + . ID=LOC_Os01g01030.1:exon_1;Parent=LOC_Os01g01030.1 Chr1 MSU_osa1r7 exon 13906 14271 . + . ID=LOC_Os01g01030.1:exon_2;Parent=LOC_Os01g01030.1 Chr1 MSU_osa1r7 exon 14359 14437 . + . ID=LOC_Os01g01030.1:exon_3;Parent=LOC_Os01g01030.1 Chr1 MSU_osa1r7 exon 14969 15171 . + . ID=LOC_Os01g01030.1:exon_4;Parent=LOC_Os01g01030.1 Chr1 MSU_osa1r7 exon 15266 15915 . + . ID=LOC_Os01g01030.1:exon_5;Parent=LOC_Os01g01030.1 Chr1 MSU_osa1r7 five_prime_UTR 12648 12773 . + . ID=LOC_Os01g01030.1:utr_1;Parent=LOC_Os01g01030.1 Chr1 MSU_osa1r7 CDS 12774 13813 . + . ID=LOC_Os01g01030.1:cds_1;Parent=LOC_Os01g01030.1 Chr1 MSU_osa1r7 CDS 13906 14271 . + . ID=LOC_Os01g01030.1:cds_2;Parent=LOC_Os01g01030.1 Chr1 MSU_osa1r7 CDS 14359 14437 . + . ID=LOC_Os01g01030.1:cds_3;Parent=LOC_Os01g01030.1 Chr1 MSU_osa1r7 CDS 14969 15171 . + . ID=LOC_Os01g01030.1:cds_4;Parent=LOC_Os01g01030.1 Chr1 MSU_osa1r7 CDS 15266 15359 . + . ID=LOC_Os01g01030.1:cds_5;Parent=LOC_Os01g01030.1 Chr1 MSU_osa1r7 three_prime_UTR 15360 15915 . + . ID=LOC_Os01g01030.1:utr_2;Parent=LOC_Os01g01030.1