Install coge: Difference between revisions
No edit summary |
No edit summary |
||
| Line 90: | Line 90: | ||
=== Deploy new CoGe Web-site === | === Deploy new CoGe Web-site === | ||
Generate an RSS key | |||
<pre>ssh-keygen | |||
cd .ssh</pre> | |||
Copy the contents of the public key into Github, and then download the CoGe repository | |||
git clone https://github.com/LyonsLab/coge.git | git clone https://github.com/LyonsLab/coge.git | ||
| Line 184: | Line 187: | ||
</pre> | </pre> | ||
<br> Configure coge.conf file | <br> Configure coge.conf file | ||
Revision as of 21:37, 10 July 2014
Installing CoGe on a blank Ubuntu machine.
Initial Dependencies
Run the following command:
sudo apt-get -y install {program}
where {program} is each of the following:
git mysql-server samtools ubuntu-dev-tools build-essential checkinstall gcc-multilib expat libexpat1-dev libgd2-xpm-dev build-essential njplot imagemagick graphviz apache2 lamp-server^ phpmyadmin swig ttf-mscorefonts-installer
Then, use:
sudo cpan App::cpanminus
sudo cpanm {module}
where {module} is each of the following:
AuthCas DBIx::Class DBIx::Class::Schema DBD::SQLite Data::UUID Data::GUID XML::SAX::Expat XML::Parser XML::Simple IPC::System::Simple CGI::Application::Dispatch CGI::Log GD::Graph::bars Moose ZMQ::LibZMQ3 File::Slurp DateTime URI::Split JSON JSON::XS
Create new mysql database
Dump CoGe Database schema
- File download: http://data.iplantcollaborative.org/quickshare/71b18508287f9fb0/cogetable.sql (AUTO_INCREMENT removed)
mysqldump -d -h localhost -u root -pXXXXXXX coge | sed 's/AUTO_INCREMENT=[0-9]*\b//' > cogetable.sql
Note: be sure to disable apparmour for MySQL.
Create new CoGe Database
create database coge
Initialize new coge database
mysql -u root -pXXXXXXXX coge < cogetable.sql
Populate a few entries in the feature_type table
- This is important because part of CoGe's code-base is keyed to feature_type_ids. This is done in order to improve performance of the system by using a feature_type_id to retrieve features of a particular type. An example is OrganismView which needs to find features of type "chromosome" in order to determine the size of a genome. The table loaded here contains 10 feature types
- Download file: http://data.iplantcollaborative.org/quickshare/ac8758f83c9b29b1/feature_type.sql
mysql -u root -pXXXXXXXXX coge < feature_type.sql
Create new user for new CoGe database
- Want a web-user with limited write privileges and a power user to load new data
create user 'coge'@'localhost' IDENTIFIED BY 'XXXXXX'; grant all privileges on coge.* to coge; create user 'coge_web'@'localhost' IDENTIFIED BY 'XXXXXX'; grant select on coge.* to coge_web; flush privileges;
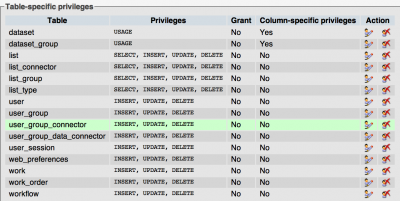
Note: The CoGe web-user needs edit/insert permission on some tables. Here is a snapshot of what these are:
Deploy new CoGe Web-site
Generate an RSS key
ssh-keygen cd .ssh
Copy the contents of the public key into Github, and then download the CoGe repository
git clone https://github.com/LyonsLab/coge.git
Make directories where files go and give write persmission
cd coge/web ./setup.sh
Configure apache
The /etc/apache2/sites-available/default.conf should look like this:
<VirtualHost *>
ServerAdmin webmaster@localhost
DocumentRoot /opt/coge/web
<Files ~ "\.(pl|cgi)$">
SetHandler perl-script
PerlResponseHandler ModPerl::Registry
Options +ExecCGI
PerlSendHeader On
</Files>
<Directory />
Options FollowSymLinks
AllowOverride None
</Directory>
<Directory /opt/coge/web>
PerlSetEnv COGE_HOME "/opt/coge/web"
Options Indexes FollowSymLinks MultiViews
AllowOverride None
Order allow,deny
allow from all
# This directive allows us to have apache2's default start page
# in /apache2-default/, but still have / go to the right place
#RedirectMatch ^/$ /apache2-default/
</Directory>
<Directory /opt/coge/web/services/>
Options +FollowSymLinks +ExecCGI
AddHandler wsgi-script .py
</Directory>
<Directory /opt/coge/web/services/JBrowse/JBrowse_TrackContent_WS/>
Options +FollowSymLinks +ExecCGI
AddHandler wsgi-script .py
</Directory>
Alias /CoGe/jex /opt/coge/web/services/jex.py
Alias /CoGe/services/JBrowse/track /opt/coge/web/services/JBrowse/J$
Alias /CoGe "/opt/coge/web"
Alias /gobe/ /opt/coge/web/gobe/
<Directory "/opt/coge/web/gobe/">
Options +FollowSymLinks +ExecCGI
AddHandler wsgi-script .py
</Directory>
ScriptALias /api/v1/jbrowse/ /opt/coge/web/services/service.pl
Alias /services/JBrowse/track /opt/coge/web/services/JBrowse/JBrowse_TrackContent_WS/source.py
ScriptAlias /api/v1/ /opt/coge/web/services/mojolicious/
ScriptAlias /cgi-bin/ /opt/coge/web/
<Directory "/opt/coge/web">
AllowOverride None
Options +ExecCGI -MultiViews +SymLinksIfOwnerMatch
AddHandler cgi-script .pl
Order allow,deny
Allow from all
</Directory>
wsgiScriptAlias /jex /opt/coge/web/services/jex.py
ErrorLog /var/log/apache2/error.log
# Possible values include: debug, info, notice, warn, error, crit,
# alert, emerg.
LogLevel warn
CustomLog /var/log/apache2/access.log combined
ServerSignature On
Alias /doc/ "/usr/share/doc/"
<Directory "/usr/share/doc/">
Options Indexes MultiViews FollowSymLinks
AllowOverride None
Order deny,allow
Deny from all
Allow from 127.0.0.0/255.0.0.0 ::1/128
</Directory>
</VirtualHost>
Configure coge.conf file
Replacing XXX's with your own information. (Change paths as necessary; this template is configured for having the Coge directory in the path: /opt/coge)
##This is a configuration file for CoGe. #database configuration DBNAME coge DBHOST localhost DBPORT 3307 DBUSER coge DBPASS XXXXXXX #CAS authentication for webservices CAS_URL https://auth.iplantcollaborative.org/cas #basic auth name and password AUTHNAME XXXXXX AUTHPASS XXXXXX #web cookie name COOKIE_NAME cogec #support email address SUPPORT_EMAIL XXXXXX #basedir for coge COGEDIR /opt/coge/web/ #bin dir for coge's programs BINDIR /opt/coge/web/bin/ #scripts dir for coge's programs SCRIPTDIR /opt/coge/scripts RESOURCESDIR /opt/coge/resources #data dir for coge's programs DATADIR /storage/coge/data/ #cache dir CACHEDIR /storage/coge/data/cache/ #dir for pair-wise whole genome comparisons (e.g. SynMap) DIAGSDIR /opt/coge/web/data/diags/ #fasta dir FASTADIR /opt/coge/web/data/fasta/ #sequence dir SEQDIR /storage/coge/data/genomic_sequence/ #SEQDIR /opt/tmp/data/ #experiment dir EXPDIR /storage/coge/data/experiments/ #TMPL dir for CoGe's web page templates TMPLDIR /opt/coge/web/tmpl/ #temp dir for coge TEMPDIR /opt/coge/web/tmp/ #secure temp dir SECTEMPDIR /storage/coge/tmp/ #IRODS dir IRODSDIR /iplant/home/<USER>/coge_data IRODSENV /opt/coge/web/irodsEnv #Base URL for web-server URL / #URL for temp directory TEMPURL /tmp/ #blast style scoring matrix dirs #BLASTMATRIX /opt/coge/web/data/blast/matrix/ BLASTMATRIX /storage/coge/data/blast/matrix/ #blastable DB #BLASTDB /opt/coge/web/data/blast/db/ BLASTDB /storage/coge/data/blast/db/ #lastable DB #LASTDB /home/franka1/coge/web/data/last/db/ LASTDB /storage/coge/data/last/db/ #directory for bed files BEDDIR /opt/coge/web/data/bed/ #servername for links SERVER http://XXXXXX/ #Job Engine Server JOBSERVER localhost #Job Engine Port JOBPORT 5151 #directory for caching genome browser images IMAGE_CACHE /opt/coge/web/data/image_cache/ #maximum number of processor to use for multi-CPU systems MAX_PROC 32 COGE_BLAST_MAX_PROC 8 #True Type Font FONT /usr/share/fonts/truetype/msttcorefonts/arial.ttf #SynMap workflow tools KSCALC /opt/coge/web/bin/SynMap/kscalc.pl GEN_FASTA /opt/coge/web/bin/SynMap/generate_fasta.pl RUN_ALIGNMENT /opt/coge/web/bin/SynMap/quota_align_merge.pl RUN_COVERAGE /opt/coge/web/bin/SynMap/quota_align_coverage.pl PROCESS_DUPS /opt/coge/web/bin/SynMap/process_dups.pl GEVO_LINKS /opt/coge/web/bin/SynMap/gevo_links.pl DOTPLOT_DOTS /opt/coge/web/bin/dotplot_dots.pl #various programs BL2SEQ /usr/local/bin/legacy_blast.pl bl2seq BLASTZ /usr/local/bin/blastz LASTZ /usr/local/bin/lastz MULTI_LASTZ /opt/coge/web/bin/blastz_wrapper/blastz.py LAST_PATH /opt/coge/web/bin/last_wrapper/ MULTI_LAST /opt/coge/web/bin/last_wrapper/last.py #BLAST 2.2.23+ BLAST /usr/local/bin/legacy_blast.pl blastall TBLASTN /usr/local/bin/tblastn BLASTN /usr/local/bin/blastn BLASTP /usr/local/bin/blastp TBLASTX /usr/local/bin/tblastx FASTBIT_LOAD /usr/local/bin/ardea FASTBIT_QUERY /usr/local/bin/ibis SAMTOOLS /usr/bin/samtools RAZIP /usr/local/bin/razip ###Formatdb needs to be updated to makeblastdb FORMATDB /usr/bin/formatdb LAGAN /opt/coge/web/bin/lagan-64bit/lagan.pl LAGANDIR /opt/coge/web/bin/lagan-64bit/ CHAOS /opt/coge/web/bin/lagan-64bit/chaos GENOMETHREADER /opt/coge/web/bin/gth DIALIGN /opt/coge/web/bin/dialign2_dir/dialign2-2_coge DIALIGN2 /opt/coge/web/bin/dialign2_dir/dialign2-2_coge DIALIGN2_DIR /opt/coge/web/bin/dialign2_dir/ HISTOGRAM /opt/coge/web/bin/histogram.pl KS_HISTOGRAM /opt/coge/web/bin/ks_histogram.pl PYTHON /usr/bin/python PYTHON26 /usr/bin/python DAG_TOOL /opt/coge/web/bin/SynMap/dag_tools.py BLAST2BED /opt/coge/web/bin/SynMap/blast2bed.pl TANDEM_FINDER /opt/coge/web/bin/dagchainer/tandems.py DAGCHAINER /opt/coge/web/bin/dagchainer_bp/dag_chainer.py EVALUE_ADJUST /opt/coge/web/bin/dagchainer_bp/dagtools/evalue_adjust.py FIND_NEARBY /opt/coge/web/bin/dagchainer_bp/dagtools/find_nearby.py QUOTA_ALIGN /opt/coge/web/bin/quota-alignment/quota_align.py CLUSTER_UTILS /opt/coge/web/bin/quota-alignment/cluster_utils.py BLAST2RAW /opt/coge/web/bin/quota-alignment/scripts/blast_to_raw.py SYNTENY_SCORE /opt/coge/web/bin/quota-alignment/scripts/synteny_score.py DOTPLOT /opt/coge/web/bin/dotplot.pl SVG_DOTPLOT /opt/coge/web/bin/SynMap/dotplot.py NWALIGN /usr/local/bin/nwalign CODEML /opt/coge/web/bin/codeml/codeml-coge CODEMLCTL /opt/coge/web/bin/codeml/codeml.ctl CONVERT_BLAST /opt/coge/web/bin/convert_long_blast_to_short_blast_names.pl DATASETGROUP2BED /opt/coge/web/bin/dataset_group_2_bed.pl ARAGORN /usr/local/bin/aragorn CLUSTALW /usr/local/bin/clustalw2 GZIP /bin/gzip GUNZIP /bin/gunzip TAR /bin/tar #MotifView MOTIF_FILE /opt/coge/web/bin/MotifView/motif_hash_dump #stuff for Mauve and whole genome alignments MAUVE /opt/coge/web/bin/GenomeAlign/progressiveMauve-muscleMatrix COGE_MAUVE /opt/coge/web/bin/GenomeAlign/mauve_alignment.pl MAUVE_MATRIX /opt/coge/web/data/blast/matrix/nt/Mauve-Matrix-GenomeAlign #newicktops is part of njplot package NEWICKTOPS /usr/bin/newicktops #convert is from ImageMagick CONVERT /usr/bin/convert #from graphviz DOT NEATO CUTADAPT /usr/local/bin/cutadapt GSNAP /usr/local/bin/gsnap CUFFLINKS /usr/local/bin/cufflinks PARSE_CUFFLINKS /opt/coge/scripts/parse_cufflinks.py GMAP_BUILD /usr/local/bin/gmap_build BOWTIE_BUILD /usr/local/bin/bowtie2-build TOPHAT /usr/local/bin/tophat #THIRD PARTY URLS GENFAMURL http://dev.gohelle.cirad.fr/genfam/?q=content/upload GRIMMURL http://grimm.ucsd.edu/cgi-bin/grimm.cgi#report QTELLER_URL http://geco.iplantc.org/qTeller
Install Perl Modules and other remaining dependencies
- Install CoGe modules /modules directory of CoGe install path
sudo perl Makefile.PL lib=/usr/local/lib/perl/5.18.2; sudo make install
- Each .pm file in /coge/web will have a list of perl modules at the top of the file. Use 'sudo cpanm' to install these.
- For each path in coge.conf that starts with /usr/local/bin, download these programs and follow the installation instructions on their respective websites.
- GMAP Gsnap, and fastbit (be sure to get version 1.3.5, rather than the most recent) require the command './configure && make && sudo make install'
- For clustalw, cufflinks, TopHat, and Bowtie (needs unzipping), copy the executable files into /usr/local/bin
- Nwalign and cutadapt need the command 'sudo python setup.py install'
Install JBrowse
Download and install the JBrowse package from http://jbrowse.org/install/
cd js unzip JBrowse-1.11.4-dev.zip mv JBrowse-1.11.4 /coge/web/js/jbrowse
Populate with test data
scripts/replicate_genome_between_coge_installations.pl -dsgid 11022 -u1 coge -p1 XXXXXXX -db1 coge -u2 oryza_coge -p2 XXXXXXX -db2 oryza_coge -sd /opt/apache/oryz_coge/data/genomic_sequence
Troubleshooting
Visualization in GEvo does not work
This relies on a system known as Gobe. Check the following things:
- Apache configuration for gobe
- Check to see if paths hard-coded into gobe/flash/service.wsgi need to be updated
- NOTE: Not sure if this is required