Syntenic dotplot: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Image:Dotplot.png|thumb|center|1000px]] | [[Image:Dotplot.png|thumb|center|1000px|Syntenic dotplot of E-coli B strain REL 606 and E-coli K12 dh 10b]] | ||
{| width="1000" cellspacing="1" cellpadding="1" border="1" | {| width="1000" cellspacing="1" cellpadding="1" border="1" | ||
|- | |- | ||
| Variation observed in B and K12<br> | | Variation observed in B and K12<br> | ||
| Description of variation<br> | | Description of variation<br> | ||
| Link leading to GEvo <br> | | Link leading to GEvo <br> | ||
|- | |- | ||
| 1. Deletion in B strain <br> | | 1. Deletion in B strain <br> | ||
| DNA pol II, genes in metabolic pathway, thiamine ABC transporter present in K12 strain Transposase in K12 strain. Prophage CP46 DNA insertion in K12 strain. <br> | | DNA pol II, genes in metabolic pathway, thiamine ABC transporter present in K12 strain Transposase in K12 strain. Prophage CP46 DNA insertion in K12 strain. <br> | ||
| http://tinyurl.com/yexrzpb<br> | | http://tinyurl.com/yexrzpb<br> | ||
|- | |- | ||
| 2. Insertions in K12 strain <br> | | 2. Insertions in K12 strain <br> | ||
| Transposase in K12 strain. Prophage CP46 DNA insertion in K12 strain. <br> | | Transposase in K12 strain. Prophage CP46 DNA insertion in K12 strain. <br> | ||
| http://tinyurl.com/yd2quy7<br> | | http://tinyurl.com/yd2quy7<br> | ||
|- | |- | ||
| 3. Deletion in B strain or Insertion in K12 strain<br> | | 3. Deletion in B strain or Insertion in K12 strain<br> | ||
| Presence of lac operon and other metabolic enzymes genes in K12 strain. Pseudogene made by transposone insertion in K12. | | Presence of lac operon and other metabolic enzymes genes in K12 strain. Pseudogene made by transposone insertion in K12. | ||
| http://tinyurl.com/yedn5tb<br> | | http://tinyurl.com/yedn5tb<br> | ||
|- | |- | ||
| 4. Insertion in B strain and DNA duplication event in K12. <br> | | 4. Insertion in B strain and DNA duplication event in K12. <br> | ||
| Prophage DNA insertion in B strain. Recent DNA duplication event in K12 as seen by 100% identity between paralogs. | | Prophage DNA insertion in B strain. Recent DNA duplication event in K12 as seen by 100% identity between paralogs. | ||
| http://tinyurl.com/yea8bu6<br> | | http://tinyurl.com/yea8bu6<br> | ||
|- | |- | ||
| 5. Insertion in B strain<br> | | 5. Insertion in B strain<br> | ||
| Bacteriophage DNA insertion in B strain | | Bacteriophage DNA insertion in B strain | ||
| http://tinyurl.com/yevlb2w<br> | | http://tinyurl.com/yevlb2w<br> | ||
|- | |- | ||
| 6. Insertion in B strain<br> | | 6. Insertion in B strain<br> | ||
| Prophage DNA insertion in B strain | | Prophage DNA insertion in B strain | ||
| http://tinyurl.com/ybokuag<br> | | http://tinyurl.com/ybokuag<br> | ||
|- | |- | ||
| 7. Insertion in K12 strain<br> | | 7. Insertion in K12 strain<br> | ||
| Prophage DNA insertion in K12 | | Prophage DNA insertion in K12 | ||
| http://tinyurl.com/yaxlh7o<br> | | http://tinyurl.com/yaxlh7o<br> | ||
|- | |- | ||
| 8. Insertion of transposon in K12 and B strain. Deletion of phenylacetic acid genes in B strain or insertion of those in K12. <br> | | 8. Insertion of transposon in K12 and B strain. Deletion of phenylacetic acid genes in B strain or insertion of those in K12. <br> | ||
| Rac prophage DNA insertion in K12. Insertion of IS3 element protein and transposase in B strain. Insertion of IS30 transposon in B strain created pseudogene. Presence of phenylacetic acid degradation genes in K12 | | Rac prophage DNA insertion in K12. Insertion of IS3 element protein and transposase in B strain. Insertion of IS30 transposon in B strain created pseudogene. Presence of phenylacetic acid degradation genes in K12 | ||
| http://tinyurl.com.ye7wcxe<br> | | http://tinyurl.com.ye7wcxe<br> | ||
|- | |- | ||
| 9. Insertion in K12 strains. <br>Flagella synthesizing genes either deleted in B strain or inserted in K12. <br><br> | | 9. Insertion in K12 strains. <br>Flagella synthesizing genes either deleted in B strain or inserted in K12. <br><br> | ||
| Presence of flagella encoding/regulatory genes in K12. IS5 sequence insertion in K12. CP4-44 prophage DNA insertion in K12. | | Presence of flagella encoding/regulatory genes in K12. IS5 sequence insertion in K12. CP4-44 prophage DNA insertion in K12. | ||
| http://tinyurl.com/ybh84x7 | | http://tinyurl.com/ybh84x7 | ||
<br> | <br> | ||
|- | |- | ||
| 10. Insertion in B strain. <br>Pili associated genes and metal resistance genes either inserted in K12 or deleted in B strain.<br><br> | | 10. Insertion in B strain. <br>Pili associated genes and metal resistance genes either inserted in K12 or deleted in B strain.<br><br> | ||
| Bacteriophage DNA insertion in B strain. <br>IS1 sequence insertion in B strain. <br>Presence of pili associated genes in K12 together with genes conferring resistance to Co and Ni.<br><br> | | Bacteriophage DNA insertion in B strain. <br>IS1 sequence insertion in B strain. <br>Presence of pili associated genes in K12 together with genes conferring resistance to Co and Ni.<br><br> | ||
| http://tinyurl.com/yammj7v<br> | | http://tinyurl.com/yammj7v<br> | ||
|- | |- | ||
| 11. Insertion in K12 strain. ParB family protein and recombinase deleted in K12. <br> | | 11. Insertion in K12 strain. ParB family protein and recombinase deleted in K12. <br> | ||
| CP4-57 prophage DNA insertion in K12 strain. Presence of ParB family protein gene and recombinase in B strain<br> | | CP4-57 prophage DNA insertion in K12 strain. Presence of ParB family protein gene and recombinase in B strain<br> | ||
| http://tinyurl.com/ycdog2c<br> | | http://tinyurl.com/ycdog2c<br> | ||
|- | |- | ||
| 12. Insertion in B and K12 strain. Inversion. Insertion of saframycin synthetase in B strain or deletion of that in K12. <br> | | 12. Insertion in B and K12 strain. Inversion. Insertion of saframycin synthetase in B strain or deletion of that in K12. <br> | ||
| Prophage DNA insertion in B strain. <br>Inversion of prepilin leader peptidase/methylase and M-type protein. Is5 insertion in K12. Presence of saframycin synthetase gene in B strain. | | Prophage DNA insertion in B strain. <br>Inversion of prepilin leader peptidase/methylase and M-type protein. Is5 insertion in K12. Presence of saframycin synthetase gene in B strain. | ||
<br> | <br> | ||
| http://tinyurl.com/ycoagxh<br> | | http://tinyurl.com/ycoagxh<br> | ||
|- | |- | ||
| 13. Insertion of transposon, ShiA-like, TrbC-like proteins in B strain. <br> | | 13. Insertion of transposon, ShiA-like, TrbC-like proteins in B strain. <br> | ||
| IS30 transposon insertion in B strain. Presence of ShiA-like and TrbC-like protein in B strain<br> | | IS30 transposon insertion in B strain. Presence of ShiA-like and TrbC-like protein in B strain<br> | ||
| http://tinyurl.com/ydkrcv8<br> | | http://tinyurl.com/ydkrcv8<br> | ||
|- | |- | ||
| 14. Insertion in B strain<br> | | 14. Insertion in B strain<br> | ||
| Insertion of fimbriae synthesis genes, multi drug transporters, Is1 protein gene in B strain.<br> | | Insertion of fimbriae synthesis genes, multi drug transporters, Is1 protein gene in B strain.<br> | ||
| http://tinyurl.com/ycwsmsl<br> | | http://tinyurl.com/ycwsmsl<br> | ||
|} | |} | ||
<br> | <br> | ||
Revision as of 05:01, 6 October 2009
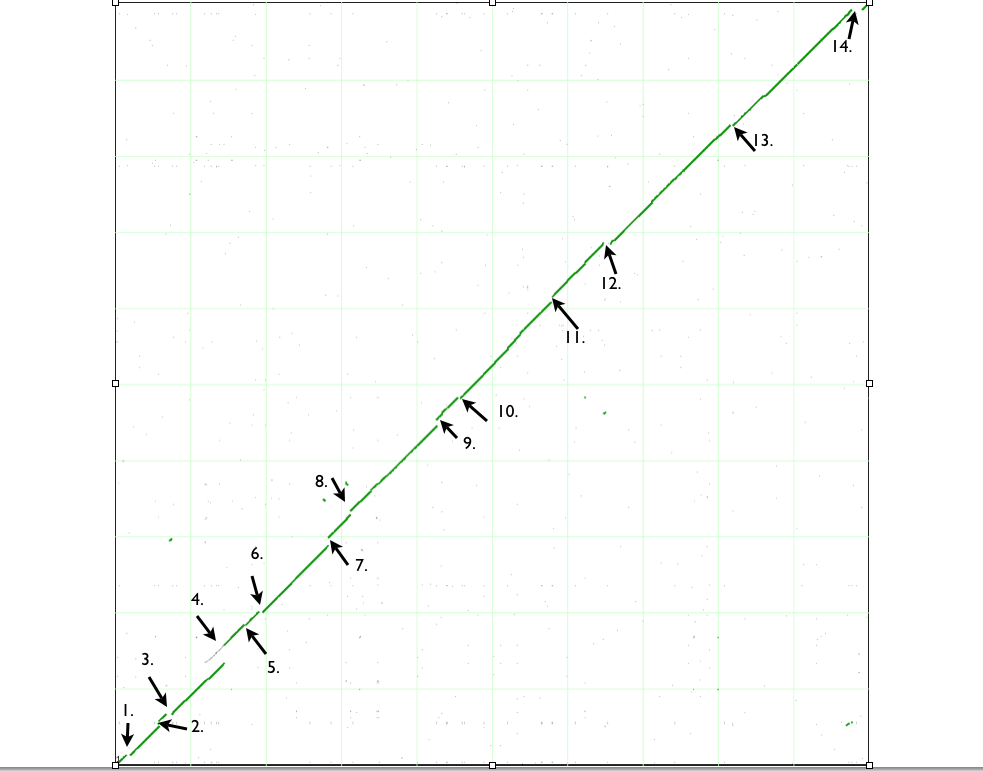
| Variation observed in B and K12 |
Description of variation |
Link leading to GEvo |
| 1. Deletion in B strain |
DNA pol II, genes in metabolic pathway, thiamine ABC transporter present in K12 strain Transposase in K12 strain. Prophage CP46 DNA insertion in K12 strain. |
http://tinyurl.com/yexrzpb |
| 2. Insertions in K12 strain |
Transposase in K12 strain. Prophage CP46 DNA insertion in K12 strain. |
http://tinyurl.com/yd2quy7 |
| 3. Deletion in B strain or Insertion in K12 strain |
Presence of lac operon and other metabolic enzymes genes in K12 strain. Pseudogene made by transposone insertion in K12. | http://tinyurl.com/yedn5tb |
| 4. Insertion in B strain and DNA duplication event in K12. |
Prophage DNA insertion in B strain. Recent DNA duplication event in K12 as seen by 100% identity between paralogs. | http://tinyurl.com/yea8bu6 |
| 5. Insertion in B strain |
Bacteriophage DNA insertion in B strain | http://tinyurl.com/yevlb2w |
| 6. Insertion in B strain |
Prophage DNA insertion in B strain | http://tinyurl.com/ybokuag |
| 7. Insertion in K12 strain |
Prophage DNA insertion in K12 | http://tinyurl.com/yaxlh7o |
| 8. Insertion of transposon in K12 and B strain. Deletion of phenylacetic acid genes in B strain or insertion of those in K12. |
Rac prophage DNA insertion in K12. Insertion of IS3 element protein and transposase in B strain. Insertion of IS30 transposon in B strain created pseudogene. Presence of phenylacetic acid degradation genes in K12 | http://tinyurl.com.ye7wcxe |
| 9. Insertion in K12 strains. Flagella synthesizing genes either deleted in B strain or inserted in K12. |
Presence of flagella encoding/regulatory genes in K12. IS5 sequence insertion in K12. CP4-44 prophage DNA insertion in K12. | http://tinyurl.com/ybh84x7
|
| 10. Insertion in B strain. Pili associated genes and metal resistance genes either inserted in K12 or deleted in B strain. |
Bacteriophage DNA insertion in B strain. IS1 sequence insertion in B strain. Presence of pili associated genes in K12 together with genes conferring resistance to Co and Ni. |
http://tinyurl.com/yammj7v |
| 11. Insertion in K12 strain. ParB family protein and recombinase deleted in K12. |
CP4-57 prophage DNA insertion in K12 strain. Presence of ParB family protein gene and recombinase in B strain |
http://tinyurl.com/ycdog2c |
| 12. Insertion in B and K12 strain. Inversion. Insertion of saframycin synthetase in B strain or deletion of that in K12. |
Prophage DNA insertion in B strain. Inversion of prepilin leader peptidase/methylase and M-type protein. Is5 insertion in K12. Presence of saframycin synthetase gene in B strain.
|
http://tinyurl.com/ycoagxh |
| 13. Insertion of transposon, ShiA-like, TrbC-like proteins in B strain. |
IS30 transposon insertion in B strain. Presence of ShiA-like and TrbC-like protein in B strain |
http://tinyurl.com/ydkrcv8 |
| 14. Insertion in B strain |
Insertion of fimbriae synthesis genes, multi drug transporters, Is1 protein gene in B strain. |
http://tinyurl.com/ycwsmsl |