Syntenic dotplot: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Image:Dotplot.png|thumb|center|983px]] | [[Image:Dotplot.png|thumb|center|983px|Syntenic dotplot between two '''Escherichia coli''' genomes: (x-axis) B strain REL606 and (y-axis) strain K-12 substrain DH10B. This dotplot was generated using [[SynMap]] and can be regenerated [http://synteny.cnr.berkeley.edu/CoGe/SynMap.pl?dsgid1=7454;dsgid2=4241;D=20;g=10;A=5;w=0;b=1;ft1=1;ft2=1;dt=geneorder;autogo=1 here].The green line (collection of dots) represent regions of similarity along the two genomes. The arrows point towards the spots where insertion or deletion has occurred in genome of either strains. The numbers correspond to the descriptions in the table below that accounts for these gaps in the dotplots.]] | ||
<br> | <br> | ||
Revision as of 00:29, 8 October 2009
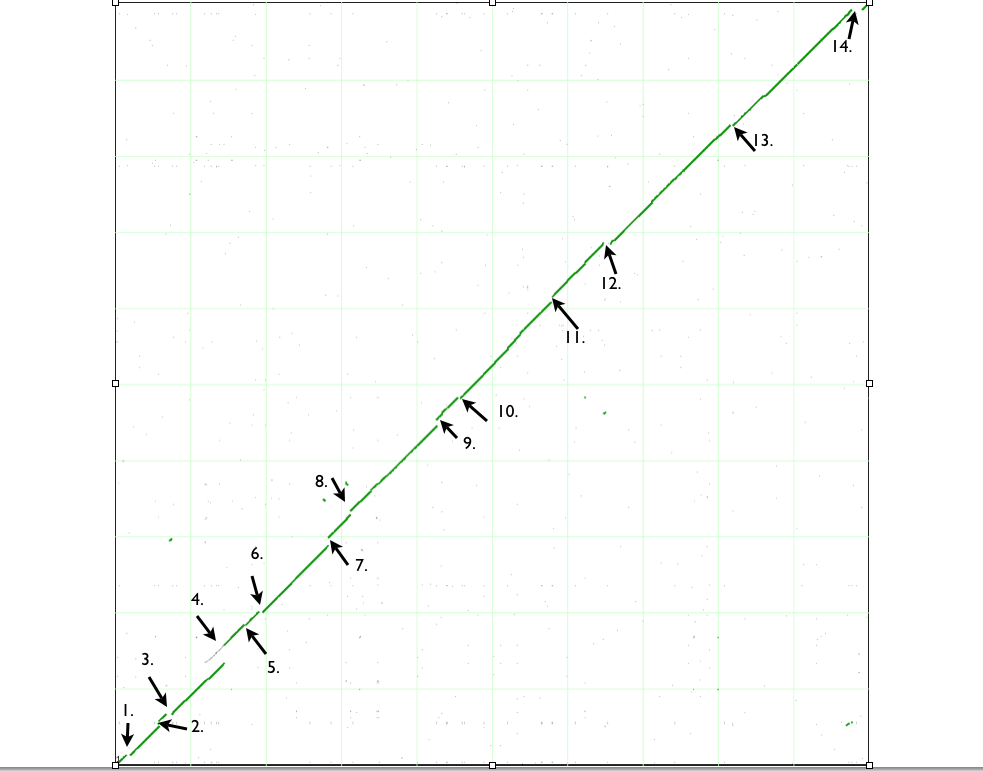
| Variation type |
Difference in strain B REL606 |
Difference in strain K-12 DH10B |
Evidence |
Notes |
Link leading to GEvo |
| 1. Deletion |
none |
Deletion of ~18 genes including DNA pol II, genes in metabolic pathway, thiamine ABC transporter |
pseudogenes in DH10B at deletion site. |
Possible additional insertion in DH10B as evidenced by pseudogenes of yabP, RNA pol associated helicase and FruR, that are not present in REl606 |
tinyurl.com/yexrzpb |
| 2. Insertion |
Insertion of IS1 sequence and a "predicted" transposase |
Insertion sequences and Prophage CP46 DNA insertion | Prophage specific genes found |
Prophage DNA insertion created pseudogenes in K-12 DH10B |
tinyurl.com/yd2quy7 |
| 3. Deletion in B strain or Insertion in K12 strain |
Insertion of IS1 sequence. Insertion of ~15 genes including lac operon and other metabolic enzymes genes | Insertion of IS3 and IS2 sequence | Pseudogene made in DH10B by IS sequence and transposon insertion |
Possible insertion in B REL606 as evidenced by direct repeats. |
http://tinyurl.com/yedn5tb |
| 4. Insertion in B strain and DNA duplication event in K12. |
Prophage DNA and transposase insertion |
Recent DNA duplication event | 100% identity between paralogs and ~98% identity between syntenic region in REL606 |
Possible phage DNA insertion in REL606 as "hypothetical protein" genes were found near putative prophage tail component gene in REL606. |
tinyurl.com/yea8bu6 |
| 5. Insertion |
Bacteriophage DNA insertion |
IS2 sequence insertion |
Pseudogenes at IS2 insertion site in DH10B. Phage specific genes were found in REL606 |
Possible phage DNA insertion in REL606 as "Hypothetical proteins" were found near phage specific genes |
tinyurl.com/yevlb2w |
| 6. Insertion |
Prophage DNA insertion |
none |
Phage specific genes were found in REL606 |
none |
tinyurl.com/ybokuag |
| 7. Insertion |
none | Prophage DNA insertion | Phage specific genes found in DH10B |
none |
tinyurl.com/yaxlh7o |
| 8. Insertion and deletion |
Insertion of IS3 element protein and deletion of phenylacetic acid degradation genes |
IS and Rac prophage DNA insertion | Phage specific genes found in DH10B. IS insertions in REL606 might have created direct repeats and facilitated excision of phenylacetic acid degradation genes. |
none | tinyurl.com/yccbmsq |
| 9. Insertion |
Deletion of flagella encoding/regulatory genes | flagella encoding/regulatory genesIS5 sequence and CP4-44 prophage DNA in DH10B. |
http://tinyurl.com/ybh84x7
| ||
| 10. Insertion in B strain. Pili associated genes and metal resistance genes either inserted in K12 or deleted in B strain. |
Bacteriophage DNA insertion in B strain. IS1 sequence insertion in B strain. Presence of pili associated genes in K12 together with genes conferring resistance to Co and Ni. |
http://tinyurl.com/yammj7v | |||
| 11. Insertion in K12 strain. ParB family protein and recombinase deleted in K12. |
CP4-57 prophage DNA insertion in K12 strain. Presence of ParB family protein gene and recombinase in B strain |
http://tinyurl.com/ycdog2c | |||
| 12. Insertion in B and K12 strain. Inversion. Insertion of saframycin synthetase in B strain or deletion of that in K12. |
Prophage DNA insertion in B strain. Inversion of prepilin leader peptidase/methylase and M-type protein. Is5 insertion in K12. Presence of saframycin synthetase gene in B strain.
|
http://tinyurl.com/ycoagxh | |||
| 13. Insertion of transposon, ShiA-like, TrbC-like proteins in B strain. |
IS30 transposon insertion in B strain. Presence of ShiA-like and TrbC-like protein in B strain |
http://tinyurl.com/ydkrcv8 | |||
| 14. Insertion in B strain |
Insertion of fimbriae synthesis genes, multi drug transporters, Is1 protein gene in B strain. |
http://tinyurl.com/ycwsmsl |