FractBias: Difference between revisions
No edit summary |
|||
| Line 149: | Line 149: | ||
''Arabidopsis'' and ''Brassica'' Fractionation Bias | ''Arabidopsis thaliana'' and ''Brassica rapa'' Fractionation Bias | ||
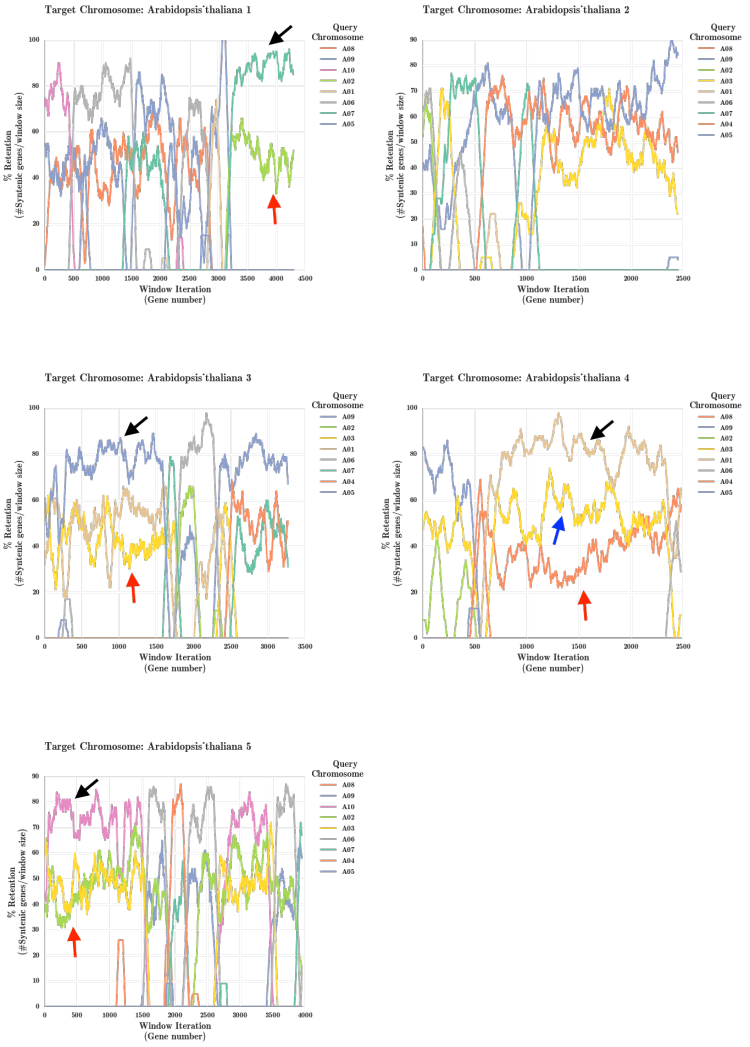
[[File:Athaliana_Brapa_onlysyntenic.png|right|thumb|750px|'''Figure 6.''' Results from running the FractBias tool ]] | |||
{| class="wikitable" | |||
!colspan="6"|'''Table 1. FractBias examples available through CoGe’s SynMap. Syntenic depth ratios range from 1:1 to 1:6 using two species of plasmodia, two mammals, and six species of plants to highlight the flexibility and ease of use of FractBias. ''' | |||
|- | |||
! scope="col"|Target Species | |||
! scope="col"|Query Species | |||
! scope="col"|Syntenic Depth Ratio | |||
! scope="col"|Link to 'All Genes' Analysis | |||
! scope="col"|Link to 'Only Syntenic Genes' Analysis | |||
|- | |||
| style="text-align:center;" |Plasmodium falciparum | |||
| style="text-align:center;" |Plasmodium knowlesi | |||
| style="text-align:center;" |1:1 | |||
| style="text-align:center;" |https://genomevolution.org/r/k7j6 | |||
| style="text-align:center;" |https://genomevolution.org/r/k7km | |||
|- | |||
| style="text-align:center;" |Homo sapiens | |||
| style="text-align:center;" |Pan troglodytes | |||
| style="text-align:center;" |1:1 | |||
| style="text-align:center;" |https://genomevolution.org/r/k813 | |||
| style="text-align:center;" |https://genomevolution.org/r/k811 | |||
|- | |||
| style="text-align:center;" |Sorghum bicolor | |||
| style="text-align:center;" |Zea mays | |||
| style="text-align:center;" |1:2 | |||
| style="text-align:center;" |https://genomevolution.org/r/k7jx | |||
| style="text-align:center;" |https://genomevolution.org/r/k7j3 | |||
|- | |||
| style="text-align:center;" |Brassica rapa | |||
| style="text-align:center;" |Brassica napus | |||
| style="text-align:center;" |1:2 | |||
| style="text-align:center;" |https://genomevolution.org/r/k7mw | |||
| style="text-align:center;" |https://genomevolution.org/r/k7k3 | |||
|- | |||
| style="text-align:center;" |Arabidopsis thaliana | |||
| style="text-align:center;" |Brassica rapa | |||
| style="text-align:center;" |1:3 | |||
| style="text-align:center;" |https://genomevolution.org/r/k7jq | |||
| style="text-align:center;" |https://genomevolution.org/r/k7jg | |||
|- | |||
| style="text-align:center;" |Vitis vinifera | |||
| style="text-align:center;" |Arabidopsis thaliana | |||
| style="text-align:center;" |1:4 | |||
| style="text-align:center;" |https://genomevolution.org/r/k7p1 | |||
| style="text-align:center;" |https://genomevolution.org/r/k7ov | |||
|- | |||
| style="text-align:center;" |Arabidopsis thaliana | |||
| style="text-align:center;" |Brassica napus | |||
| style="text-align:center;" |1:6 | |||
| style="text-align:center;" |https://genomevolution.org/r/k7qz | |||
| style="text-align:center;" |https://genomevolution.org/r/k7r6 | |||
|} | |||
==References== | ==References== | ||
{{reflist}} | {{reflist}} | ||
Revision as of 19:53, 20 April 2016
Background
Whole genome duplications (WGDs) and genome fractionation are covered more thoroughly in other CoGepedia entries. In short, WGDs create two or more copies of a genome: which are referred to as subgenomes. The duplicate subgenomes then undergo gene loss in a process called fractionation which is part of returning to a diploid state, diploidization. All things being equal, one may assume that fractionation would occur randomly across the redundant genes created after a WGD, however bias towards gene loss on one genome, called fractionation bias, has been observed in several species including: maize [1], Brassica rapa [2], and rainbow trout [3].
The FractBias code and an example data set can be found on GitHub https://github.com/bjoyce3/SynMapFractBiasAnalysis
Overview
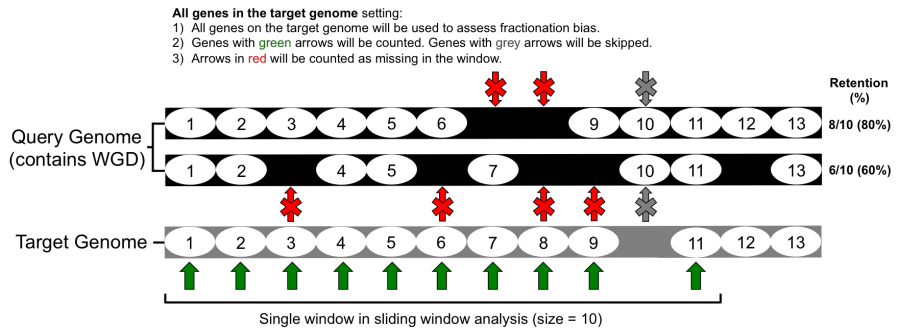
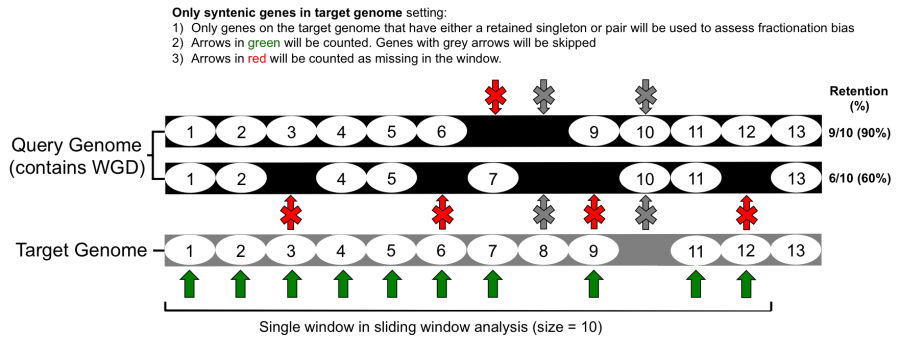
What goes in
- Two assembled genomes that have annotated coding sequences (CDS)
- A syntenic ratio set by the user (identified by empiric tests outside of the FractBias tool)
- The genome with a lower ratio will be the target genome
- The genome with a higher ratio will be the query genome
- The full GFF of the target genome
- The syntenic blocks identified by SynMap
- Setting defined by the user
- What genes should be counted
- Count all genes present on the target genome (refer to Figure 1)
- Only count genes that are retained in both genomes (refer to Figure 2)
- Target chromosome number
- Query chromosome number
- Window size
- What genes should be counted
What comes out
- A figure containing a subplot for every target genome chromosome
- Links to the raw data used to create the subplots
FractBias Methods
FractBias is a tool used to assess fractionation bias after whole genome duplications (WGDs). To investigate fractionation bias, select an organism that has experienced a WGD (e.g. maize) which will become the 'query' genome, and an organism that diverged before the WGD (e.g. sorghum recently diverged before the WGD in maize). The following is a list of all user inputs:
User Input
- Select two genomes to compare in the SynMap tool.
- Select the SynMap 'Syntenic Depth' option under 'Analysis Options.'
- Set syntenic depth ratio between genomes (determined by empirically outside of this tool).
- Set how many target genome chromosomes should be included in the analysis. There is a maximum of 40 target chromosomes that can be included, and the longest chromosomes are selected first.
- Set how many query genome chromosomes should be included in the analysis. There is a maximum of 40 query chromosomes that can be included, and the longest chromosomes are selected first.
- Set the size of the sliding window during analysis.
FractBias tool analysis
Once all of the user input options are filled and submitted, the FractBias tool then runs an analysis in the following steps:
- The coordinates for syntenic regions between the genomes are determined by the SynMap tool
- The syntenic genes are then parsed according to the 'target' and 'query' genomes. The genome with the lower syntenic depth ratio is set as the target genome; the genome with the higher ratio is set as the query genome.
- A list of genes present on every target genome chromosome is made and ordered according to start site (bp) in the annotation (gff/gtf file).
- The FractBias tool then goes through the list of each target genome gene, and determines if it has a retained homolog on one (or more) of the query chromosomes.
- Finally, the FractBias tool runs a sliding window analysis to calculate how many genes are retained for each query chromosome.
- A figure is generated that contains a subplot for every target genome chromosome
- The x-axis: target genome gene order number in sliding window analysis according to order of start site in genome annotation (gff/gtf).
- The y-axis: percent of retained genes from the target genome present on each query chromosome within that window.
- The SynMap raw data, the FractBias data, genes identified using FractBias, and the images can be downloaded through links for further use.
Code Details
There are two versions of the FractBias code written in Python 2.7.10 available on Github:
- A 'Geco' version that is run by the CoGe platform when requested through SynMap.
- An iPython version that can be run using iPython notebook
Code Assumptions and Dependencies
- The SynMap Syntenic Depth is set in Analysis Options
- The two genomes that are compared have been annotated
Code Keywords
Data Structures
- d{} is the dictionary built
Code Explanation
Data files passed in
- SynMap DAGChainer output: comparison_name.aligncoords.gcoords
- GFF file for target genome
Example Output
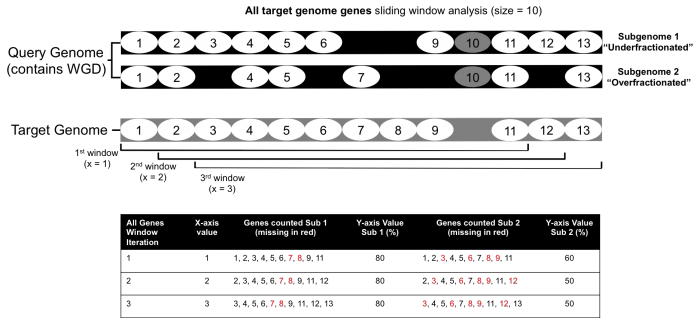
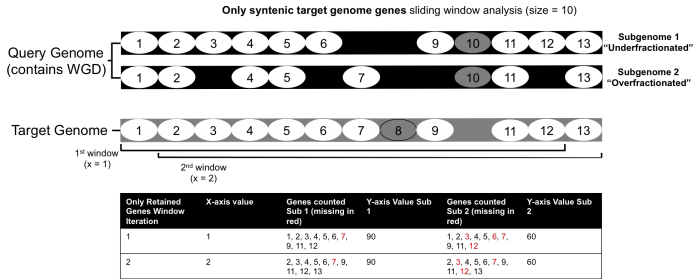
To demonstrate how the FractBias tool works, an example of a single syntenic block with eight genes is presented. The FractBias tool can be run with either "all genes" included, or "only retained genes" included. The "all genes" option will include all the genes from the target genome
| Table 1. FractBias examples available through CoGe’s SynMap. Syntenic depth ratios range from 1:1 to 1:6 using two species of plasmodia, two mammals, and six species of plants to highlight the flexibility and ease of use of FractBias. | |||||
|---|---|---|---|---|---|
| Target Species | Query Species | Syntenic Depth Ratio | Link to 'All Genes' Analysis | Link to 'Only Syntenic Genes' Analysis | |
| Plasmodium falciparum | Plasmodium knowlesi | 1:1 | https://genomevolution.org/r/k7j6 | https://genomevolution.org/r/k7km | |
| Homo sapiens | Pan troglodytes | 1:1 | https://genomevolution.org/r/k813 | https://genomevolution.org/r/k811 | |
| Sorghum bicolor | Zea mays | 1:2 | https://genomevolution.org/r/k7jx | https://genomevolution.org/r/k7j3 | |
| Brassica rapa | Brassica napus | 1:2 | https://genomevolution.org/r/k7mw | https://genomevolution.org/r/k7k3 | |
| Arabidopsis thaliana | Brassica rapa | 1:3 | https://genomevolution.org/r/k7jq | https://genomevolution.org/r/k7jg | |
| Vitis vinifera | Arabidopsis thaliana | 1:4 | https://genomevolution.org/r/k7p1 | https://genomevolution.org/r/k7ov | |
| Arabidopsis thaliana | Brassica napus | 1:6 | https://genomevolution.org/r/k7qz | https://genomevolution.org/r/k7r6 | |
If the include "only retained genes" option is set, all unique genes from either the target or query genome are not considered for the fractionation bias analysis. This option can be used to remove variation from two genomes that have diverged over longer periods and clean up the analysis.
Biological Examples
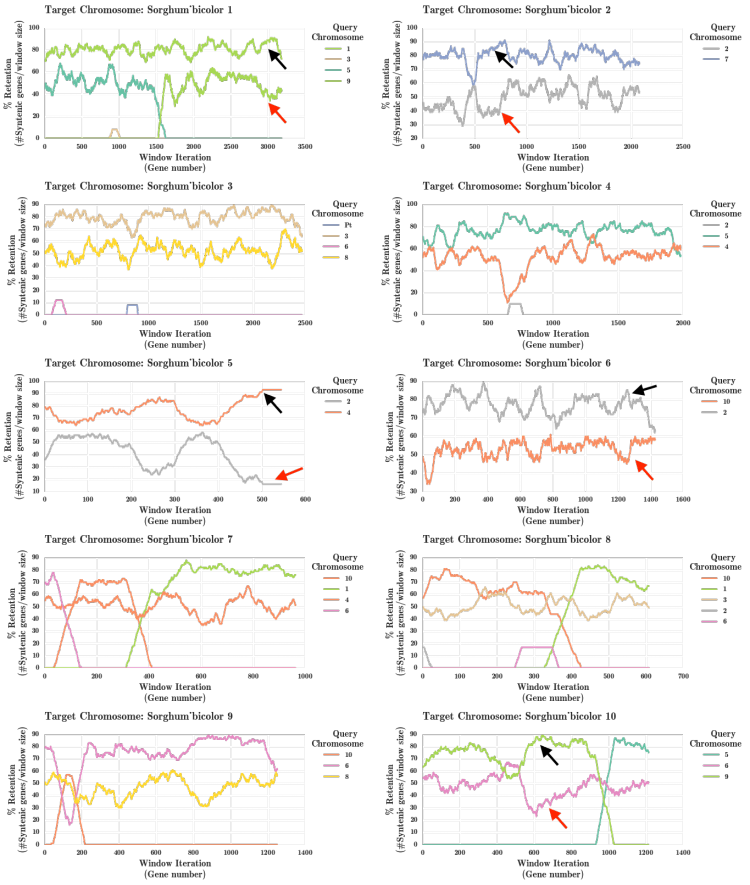
Sorghum and Maize Fractionation Bias
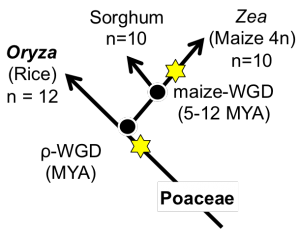
The fractionation bias in the maize genome has been previously studied[4] independently. This analysis was rerun using the FractBias tool.
Arabidopsis thaliana and Brassica rapa Fractionation Bias
| Table 1. FractBias examples available through CoGe’s SynMap. Syntenic depth ratios range from 1:1 to 1:6 using two species of plasmodia, two mammals, and six species of plants to highlight the flexibility and ease of use of FractBias. | |||||
|---|---|---|---|---|---|
| Target Species | Query Species | Syntenic Depth Ratio | Link to 'All Genes' Analysis | Link to 'Only Syntenic Genes' Analysis | |
| Plasmodium falciparum | Plasmodium knowlesi | 1:1 | https://genomevolution.org/r/k7j6 | https://genomevolution.org/r/k7km | |
| Homo sapiens | Pan troglodytes | 1:1 | https://genomevolution.org/r/k813 | https://genomevolution.org/r/k811 | |
| Sorghum bicolor | Zea mays | 1:2 | https://genomevolution.org/r/k7jx | https://genomevolution.org/r/k7j3 | |
| Brassica rapa | Brassica napus | 1:2 | https://genomevolution.org/r/k7mw | https://genomevolution.org/r/k7k3 | |
| Arabidopsis thaliana | Brassica rapa | 1:3 | https://genomevolution.org/r/k7jq | https://genomevolution.org/r/k7jg | |
| Vitis vinifera | Arabidopsis thaliana | 1:4 | https://genomevolution.org/r/k7p1 | https://genomevolution.org/r/k7ov | |
| Arabidopsis thaliana | Brassica napus | 1:6 | https://genomevolution.org/r/k7qz | https://genomevolution.org/r/k7r6 | |
References
- ↑ Schnable, J.C. et al. Dose–sensitivity, conserved non-coding sequences, and duplicate gene retention through multiple tetraploidies in the grasses. Front. Plant Sci. http://dx.doi.org/10.3389/fpls.2011.00002 (2011)
- ↑ Cheng, F. et al. Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLOS ONE DOI: 10.1371/journal.pone.0036442 (2012)
- ↑ Berthelot, C. et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nature Communications 5: DOI:10.1038/ncomms4657 (2014)
- ↑ 4.0 4.1 Schnable, J. C. et al. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. PNAS 108:4069-4074