How to load genomes into CoGe: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 9: | Line 9: | ||
## Once your genome is loaded into the system, you can add annotation and quantitative data to it using the [[LoadAnnotation]] and [[LoadExperiment]] features. | ## Once your genome is loaded into the system, you can add annotation and quantitative data to it using the [[LoadAnnotation]] and [[LoadExperiment]] features. | ||
#'''Note:''' Make sure your GFF file is in the correct format for CoGe: [[GFF ingestion]] | #'''Note:''' Make sure your GFF file is in the correct format for CoGe: [[GFF ingestion]] | ||
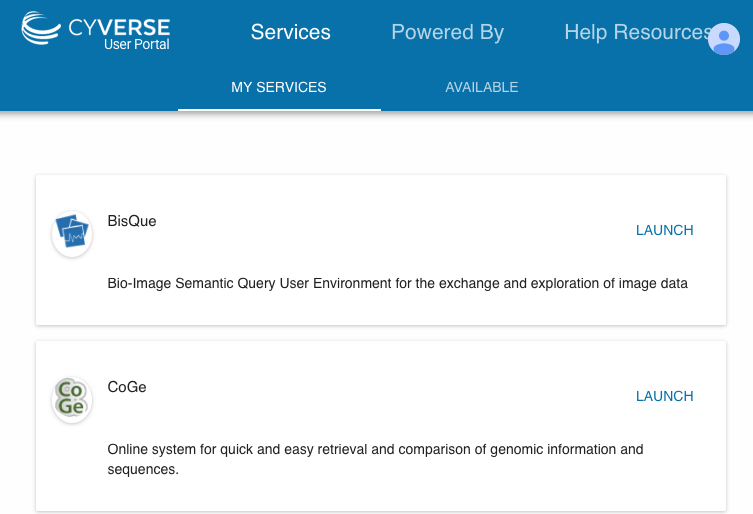
#If you get an error when accessing the CyVerse Data Store, make sure that CoGe has been added as a service to your CyVerse account: https://user.cyverse.org | |||
## [[File:Screen Shot 2017-08-14 at 3.41.40 PM.png]] | |||
Revision as of 22:42, 14 August 2017
You can load your own genome sequence, annotation, and quantitative data for use with CoGe's tools. These data may be kept private and shared with collaborators, or made fully public.
- Log into CoGe
- Go to your User Profile Page by clicking My Profile in the menu bar.

- Your profile page will look something like:
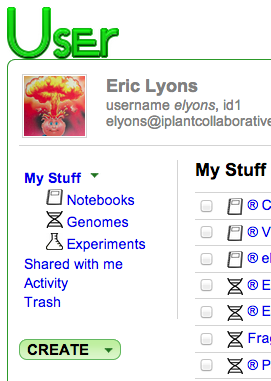
CoGe's My Profile Page - Click Create -> Load Genome
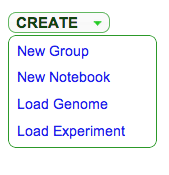
- Follow this link for information on how to use LoadGenome.
- Once your genome is loaded into the system, you can add annotation and quantitative data to it using the LoadAnnotation and LoadExperiment features.
- Note: Make sure your GFF file is in the correct format for CoGe: GFF ingestion
- If you get an error when accessing the CyVerse Data Store, make sure that CoGe has been added as a service to your CyVerse account: https://user.cyverse.org