Open-ended Analysis Network
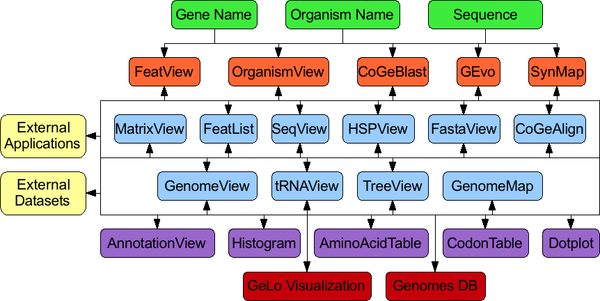
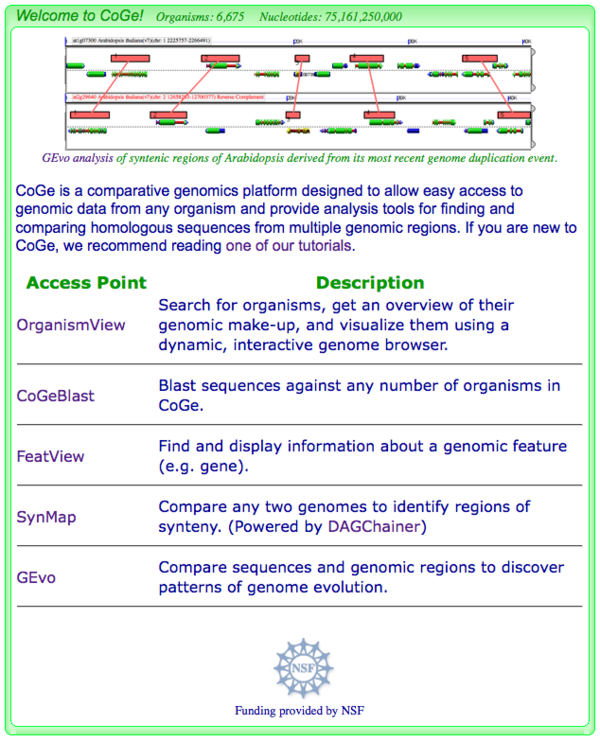
CoGe's web-tools have several applications for initiating an analysis.
- OrganismView: Search for an organism by name or description
- CoGeBlast: Blast a sequence against any number of genomes
- FeatView: Search for a genomic feature (e.g. gene) by name
- SynMap: Generate syntenic dotplots for any two genomes
- GEvo: Compare any number of genomic regions using a variety of sequence comparison algorithms
Behind the scenes, CoGe has several interconnected web-tools. Since each tool performs a specific task (e.g. displaying a list of features, viewing a fasta sequence), tools are linked together to allow researches the ability to move through their analyses in a variety of ways. This allows scientific questions to drive the analysis as opposed to the tools driving the scientific questions. Also, as new things are discovered in an analysis giving rise to new questions, each hypothesis can be immediately tested by branching an analysis down a different path than originally intended.
Also, by using a web-based framework for CoGe's tool design, it is relatively easy:
- to create new tools and plug them into the analysis network
- for other programs to link into the CoGe system and specify data to pre-load into an analysis
- to create large datasets using automated computational methods and build links to CoGe for manual proofing of data