MaizeGDB and CoGe
Overview
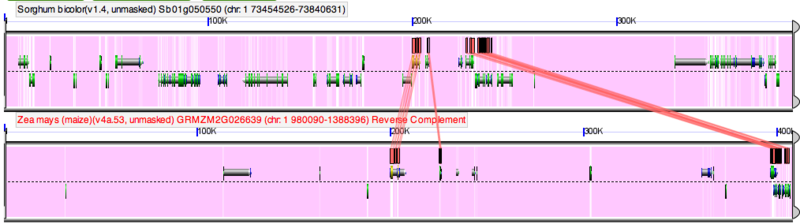
MaizeGDB is linked to CoGe's through its genome browser. A link to CoGe will be shown at the top of the browser when viewing a genomic region from the maize genome assembly (i.e. pseudomolecules). This link will take you to the same region in CoGe's GenomeView. GenomeView will automatically find maize genes with annotations that link to GEvo for syntenic gene sets with sorghum, and show those genes with special glyphs. When you view the annotations for these glyphs (by clicking on them), there will be a link to GEvo. When clicked on, these GEvo links will launch GEvo with the syntenic maize-sorghum regions pre-loaded and automatically start a GEvo analysis using blastz.
MaizeGDB's genome browser link to CoGe's genome broswer, GenomeView

MaizeGDB is one of the leading web resources for accessing genomic information about maize. Information about a maize gene or locus has links to their genome browser (powered by GBrowse of the GMOD project). At the top of the page for browsing the maize genome, is a link to CoGe's genome browser, GenomeView. If you click this link, GenomeView will open at the same genomic region.
GenomeView showing glyphs for genes linked to GEvo
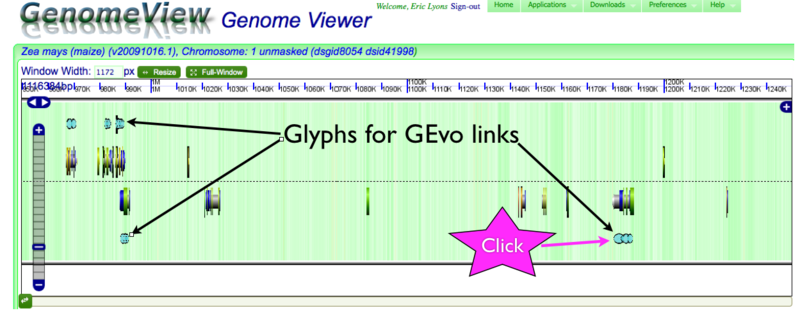
GenomeView can display a variety of genomic information. Here, the background of the virtual chromosome is colored green for GC rich DNA content and white for AT rich DNA content. The dashed line in the middle separates the top and bottoms strands of the genomic DNA duplex. Gene models are drawn as composite colored arrows above and below this line if they are read from the top and bottom strand of the genome respectively. For information about gene models and various visualization in GenomeView, please see this page. Genes with annotations that link to GEvo are displayed above and below the gene models as interlinking blue circles. To get the annotation for any gene or GEvo link, just click on it in GenomeView and a dialog box of its annotations will appear.
Getting a link to GEvo

Automatically running GEvo with syntenic maize-sorghum regions pre-loaded

GEvo's results