MaizeGDB and CoGe
Overview
MaizeGDB's genome bowers can now to used to quickly view the same region in CoGe. A link to CoGe is shown at the top of the browser when viewing a genomic region from the maize genome assembly (i.e. pseudomolecules) called "View This Region in CoGe. This link will take you to the same region in CoGe's GenomeView (in a new window so you won't lose your region at MaizeGDB). When accessed from MaizeGDB GenomeView will, by default, display the working gene set models from MaizeSequence.org release 4a.53 on B73 version 1 reference sequence. GenomeView will also display special icons to draw your attention to maize genes that have been identified as having a sorghum ortholog. Clicking on either these icons (currently a series of overlapping blue ovals) or the gene models themselves will display detailed information on the gene including the name of its sorghum ortholog and whether we've identified another maize gene as a retained homeolog from the maize whole genome duplication. The annotation window also has a link to automatically compare the orthologous genes and the syntenic regions surrounding them in another CoGe tool called GEvo.
MaizeGDB's genome browser link to CoGe's genome broswer, GenomeView

MaizeGDB is one of the leading web resources for maize genomic information. Information about a maize gene or locus has links to their genome browser (powered by GBrowse of the GMOD project) (Eric, I don't understand this sentence -James). At the top of the MaizeGDB genome browser is a link to CoGe's genome browser, GenomeView. If you click this link, GenomeView will open in a new window displaying the same genomic region.
GenomeView showing icons marking genes with identified sorghum orthologs
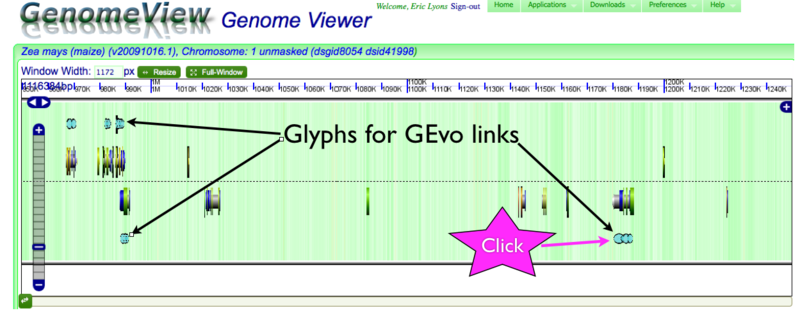
GenomeView can display a variety of genomic information. With the current default settings, show above, the background is colored green in GC rich regions and white for AT rich regions. The dashed line in the middle separates the top and bottoms strands of the DNA sequence being displayed. Gene models and other sequence features on the forward stand are drawn above the line and while gene models on the reserve strand are drawn facing the opposite direction below the line. For information about gene models and various visualizations available GenomeView, please see this page. Genes with annotations that link to GEvo are marked with special icons, currently a series of overlapping blue ovals. The current annotation information for any gene can be accessed by simply clicking on the gene model which with bring up the dialog box with annotation information and, in the case of genes marked as having sorghum orthologs, a GEvo link to compare the genes and their surrounding regions.
Getting a link to GEvo

The dialog box for an annotation in GenomeView will have a line called "GEvo link" and a description of the link to GEvo. Just click on the description of the GEvo link to launch GEvo.
Automatically running GEvo with syntenic maize-sorghum regions pre-loaded

When GEvo is launched, it will automatically have the maize-sorghum syntenic regions pre-loaded using ~400kb of the genome. In addition, it will automatically set the sequences to have all the non-protein coding sequence masked and automatically begin the analysis. Non-protein coding sequences are masked due to the highly repetitive nature of the maize genome. Having many repetitive sequences substantially increases the amount of time it takes for a GEvo analysis to process, the amount of time to render the resulting images, and the difficulty in understanding the results. By default, GEvo will use blastz for comparing these sequences. Blastz is a good algorithm to use when identifying large blocks of similar sequences.
- For information on different alignment algorithms, please see this page.
- For information on how to link to GEvo and set various parameters from the URL, please see this page.
GEvo's results
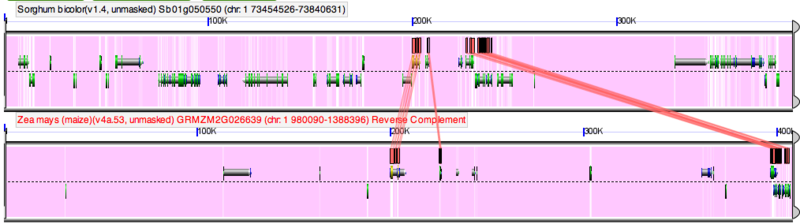
When the results from GEvo are returned, you will a panel for each of the genomic regions returned. These images are interpreted similarly as to those in GenomeView, however you will notice that the background of the chromosomes will have a lot of purple. This represents genomic sequence that was masked from the analysis (in this case, non-protein coding sequence). Also, there will be colored blocks located above and below the gene models. These blocks are regions of sequence similarity identified for pairwise comparisons of the genomic regions. If you click on one of these blocks a transparent wedge will be drawn connecting it to its partner region. By clicking on many of these (or holding the 'shift' key then click on one to connect all of them from a given track), you can detect various patterns of genome evolution.
More help and examples
If you are interested in:
- Whole genome comparisons of maize and sorghum
- Identifying syntenic regions
- Differentiating syntenic regions derived from the:
- maize-specific whole genome duplication event
- the divergence of the maize and sorghum lineages
- the pre-grass whole genome duplication event
- Identifying patterns of genome fractionation
- Identifying conserved non-coding sequences
Please see this page.
Thanks
Thanks to:
- James Schnable in Michael Freeling's lab at UC Berkeley for generating maize-sorghum syntenic gene sets.
- Taner Sen at the USDA-ARS, Iowa State University for linking MaizeGDB to CoGe and other MaizeGDB specific activities.
- Michael Freeling at UC Berkeley
- Carolyn Lawrence at USAD-ARS and Iowa State Univerity