Syntenic dotplot
Syntenic dotplots are a type of scatter-plot. Each axis represents a sequence laid end-to-end, and each dot in the scatter-plot represents a putative Homologous match between the two sequences. Often, these dotplots are used for whole genome comparisons within the same genome or across two genomes from different taxa in order to identify Synteny. Synteny is defined as two or more genomic regions that are derived from a common ancestral genomic region. The evidence for synteny is the identification of a set of genes in each genome that have a collinear arrangement. When such a pattern of gene-order conservation is discovered, the most parsimonious explanation is that the two regions are related through a common ancestor. While syntenic dotplots are useful for identifying related genomic regions, they are also useful for identifying genomic regions that have undergone an evolutionary change in one of the two genomes being compared. For example, insertions, deletions, duplications, and inversions are readily identifiable from these plots.
Below is an example of comparing two closely related substrains of E. coli strain K12. While their entire genome is highly similar to one another at the nucleotide identity level, there are many "breaks" in the syntenic path through their genomes which reveal a variety of genomic changes (mostly insertions and deletions for this example).
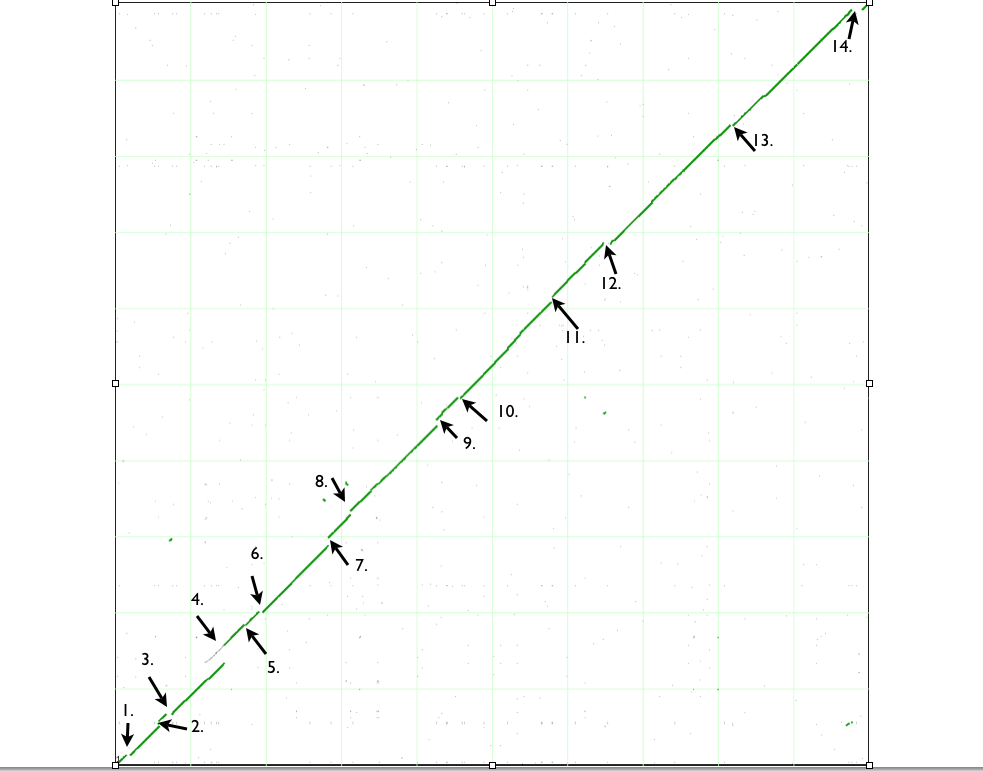
| Variation type |
Difference in strain B REL606 |
Difference in strain K-12 DH10B |
Evidence |
Notes |
Link leading to GEvo |
| 1. Deletion |
none |
Deletion of ~18 genes including DNA pol II, genes in metabolic pathway, thiamine ABC transporter |
pseudogenes in DH10B at deletion site. |
Possible additional insertion in DH10B as evidenced by pseudogenes of yabP, RNA pol associated helicase and FruR, that are not present in REl606 |
tinyurl.com/yexrzpb |
| 2. Insertion |
Insertion of IS1 transposon |
Insertion sequences and Prophage CP46 DNA insertion | Prophage specific genes found in DH10B |
Prophage DNA insertion and IS insertions has created pseudogenes in K-12 DH10B |
tinyurl.com/yd2quy7 |
| 3. Translocation in REL606 and insertion in DH10B |
Insertion of IS1 sequence. Translocation of ~15 genes including lac operon and other metabolic enzymes genes | Insertion of IS3 and IS2 sequences |
Translocation in REL606 as evidenced by direct repeats.Dotplot shows that the missing genes are present in DH10B but not in this locus. The syntenic region is therefore not colinear. |
Pseudogenes of yaiT and yaiX were created in DH10B by transposon insertions. Insertion by translocation in REL606 was confirmed as lac operon and other metabolic genes were found in DH10B by analyzing the translocated genes on the dotplot |
http://tinyurl.com/yldc83u |
| 4. Insertion in REL606 and DNA duplication event in DH10B. |
Prophage DNA and transposase insertion |
Recent DNA duplication event | 100% identity between paralogs in DH10B and ~98% identity between syntenic region of DH10B and REL606 |
Possible phage DNA insertion in REL606 as "hypothetical protein" genes were found near putative prophage tail component gene in REL606. |
tinyurl.com/yea8bu6 |
| 5. Insertion |
Bacteriophage DNA insertion |
IS2 sequence insertion |
Pseudogenes at IS2 insertion site in DH10B. Phage specific genes were found in REL606 |
Possible phage DNA insertion in REL606 as "Hypothetical proteins" were found near phage specific genes |
tinyurl.com/yevlb2w |
| 6. Insertion |
Prophage DNA insertion |
none |
Phage specific genes were found in REL606 |
none |
tinyurl.com/ybokuag |
| 7. Insertion,translocation and inversion |
none | Prophage DNA insertion and translocation of nitrite reductase 2 genes | Phage specific genes found in DH10B |
Translocation in DH10B is evident by dotplot. Moreover, the translocated genes in DH10B were found to be inverted. It could not be determined genes on which genomes were inverted as tranposon insertions were found in both genomes. |
|
| 8. Insertion and deletion |
Transposon insertions and deletion of phenylacetic acid degradation genes |
IS and Rac prophage DNA insertion | Phage specific genes found in DH10B. IS or transposon insertions in REL606 might have created direct repeats and facilitated excision of phenylacetic acid degradation genes. |
Rac prophage DNA disrupted by transposon insertion in DH10B | tinyurl.com/yccbmsq |
| 9a. Insertion | 9a. none | 9a. Insertion of IS5 sequence | 9a. none | 9a. none | 9a. http://tinyurl.com/ylllc6u |
| 9b. Insertion | 9b. Insertion of ISI transposon | 9b. none | 9b. none | 9b. none | 9b. tinyurl.com/ygsqg2f |
| 9c. Insertion | 9c. ISI insertion |
9c. Insertion of ABC transporter, flagella encoding genes and few other enzymes | 9c. Inserted DNA segment in DH10B is bordered by direct repeats at both ends. 100% identity was found between the two repeats. |
9c.DR indicates transposon insertion in DH10B. | 9ctinyurl.com/yza4jy3[1] |
| 9d. Insertion | 9d. IS2 insertion |
9d. none | 9d. none | 9d. none | 9d. tinyurl.com/ygfgtqy |
| 10. Insertion and deletion |
Bacteriophage DNA insertion and IS1 transposon insertion. Deletion of ~5 genes |
Insertion of IS3 |
IS1 insertion at the site of deletion. Another IS1 insertion might have created direct repeats and facilitated deletion. | none | tinyurl.com/ykynub2 |
| 11. Insertion and deletion |
IS1 insertion |
CP4-57 prophage DNA insertion and possible deletion of ParB family protein and recombinase |
Phage insertion in DH10B may have created pseudogene of ParB family protein genes and recombinase which later got deleted |
Pseudogenes of yqa, yga and ypj indicated possible formation of pseudogenes of ParB and recombinase at some time prior to their deletion in DH10B. |
tinyurl.com/yg7ybg4 |
| 12. Insertion, deletion and Inversion |
IS1 insertion. | IS5 and IS10 transposon insertion. Inversion of ornithine decarboxylase, M-type protein and bifunctional prepilin peptidase/methylase. Deletion of saframycin synthetase, capsule related genes, bio-film formation genes, anti-toxin system and type II secretory apparatus genes. |
Inversion in DH10B as evidenced by inverted repeats of IS10 transposon. Deletion of several genes in DH10B is evidenced by IS5 trans-activator transposase and presence of pseudogenes in DH10B, |
Insertion of IS5 trans-activator transposase indicates possible deletion of several genes in DH10B. Also, no evidence of insertion in REL606 was found such as direct repeats. |
tinyurl.com/yhyxgrq |
|
13a. Deletion |
13a. none |
13a. Deletion of putative adhesin |
13a. No direct repeats were found to indicate insertion of putative adhensin in REL606 therefore deletion in DH10B may have happened | 13a.none |
13a. tinyurl.com/yjojy53 |
|
13b. Insertion |
13b. IS1 insertion and deletion of lipopolysaccharide genes |
13b. none |
|
13b. IS1 insertion created pseudogene. | 13b.http://tinyurl.com/yj2yg5s |
|
13c. Insertion and deletion |
13c. Insertion of IS30 transposon and several 'hypothetical protein" genes. |
13c. none |
13c. Insertion in REL606 is evidenced by direct repeats
|
13c. direct repeats were found in REL606 which indicates insertion of ShiA-like and TrbC-like genes. |
13c. tinyurl.com/yjzdyum |
|
14a. Insertion |
14a. Insertion of several transposons and secondary glycine betaine transporter | 14a.Insertion of several transposons. Insertion of Kple2 phage-like element | 14a. Direct repeats bordering secondary glycine betaine transporter indicates its insertion | 14a. none | 14a. tinyurl.com/yzyvunx |
| 14b. Insertion | 14b. Insertion of ~15 genes | 14b. Phage insertion. Transposon insertions | 14b. Insertion in REL606 is evidenced by direct repeats flanking the DNA segment containing several genes. | 14b.Phage-like genes were found in DH10B | 14b. tinyurl.com/yly2b6u |
|
|
14c. none |
14c. Deletion of ~15 genes.
|
14c. Deletion in DH10B is evidenced by insertion of IS10R which may have facilitated excision of DNA by forming direct repeats |
14c. Pseudogenes found at the site of deletion and IS10R insertion. |
14c. tinyurl.com/yfhhsk6 |