CoGeBlast
 | |
 CoGeBlast Screenshot | |
| Software company | CoGe Team |
|---|---|
| Analysis Type | Blast query sequences against genomes stored in CoGe database |
| Working state | Released |
| Tools Utilized | blastn, tblastn, tblastx, blastz |
| Website | http://synteny.cnr.berkeley.edu/CoGe/CoGeBlast.pl |
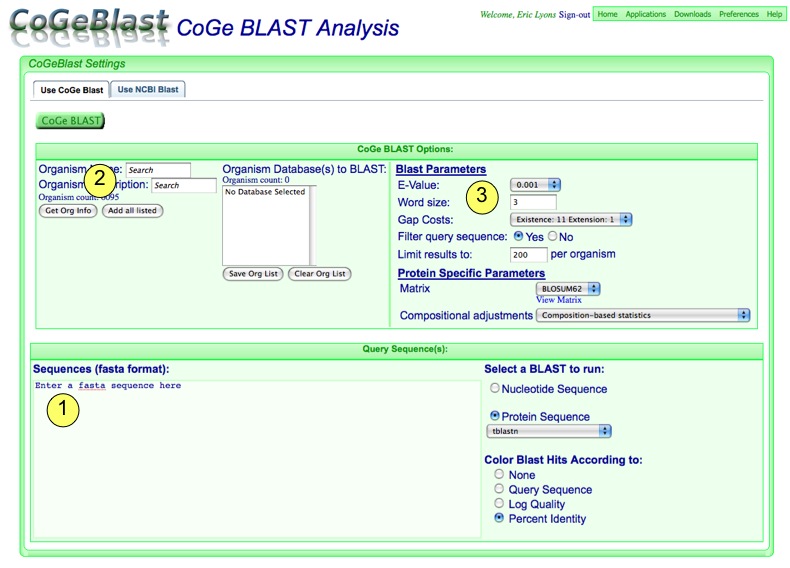
CoGeBlast is CoGe's interface to BLAST (Basic Local Alignment Search Tool) and other related algorithms. With CoGeBlast, one can take any query sequence, whether user submitted or requested from the CoGe database, and compare it against any number of genomes in the CoGe database
Overview
Background
CoGeBlast is a web-based interface to blast that allows you to quickly:
- Add query sequences
- Find and select any number of organisms and genomes in CoGe to search against
- Configure a blast analysis
- Visualize an overview of blast hits (High-scoring Sequence Pairs; HSP) in relationship to their genomic locations
- Interact with a sortable list of blast hits detailing
- which query sequence matched which organism
- their genomic location
- query sequence coverage
- variety of blast hit metrics (length, e-value, score, percent ID, quality)
- and allows you to find the closest genomic feature in the searched organisms
- Visualize individual hits in their genomic context to determine the extent to which you query matched
- Get sequence, alignment and positional information for a given hit
- An overview of the number of times your query sequences matched a given organism
- Links to data files including
- table of blast hits metrics and sequences
- fasta file of query Hit sequences
- fasta file of subject Hit sequences
- file of blast hit alignments
- raw blast reports for each organisms searched
- Ability to select identified genomic regions and:
- send to other programs in CoGe
- generate a fasta file of nearby genomic features
- export results to a tab delimited file or Microsoft Excel
Configure Screen
Alignment Algorithms
CoGeBlast utilizes a number of variants of the BLAST algorithm originally developed by Altschul et al. [1]
Documentation
You should read this if you are confused.
References
- ↑ Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990). "Basic local alignment search tool". J Mol Biol 215 (3): 403–410.doi:. PMID 2231712. http://www-math.mit.edu/~lippert/18.417/papers/altschuletal1990.pdf.
Tutorials
We has them.