Plasmodia comparative genomics

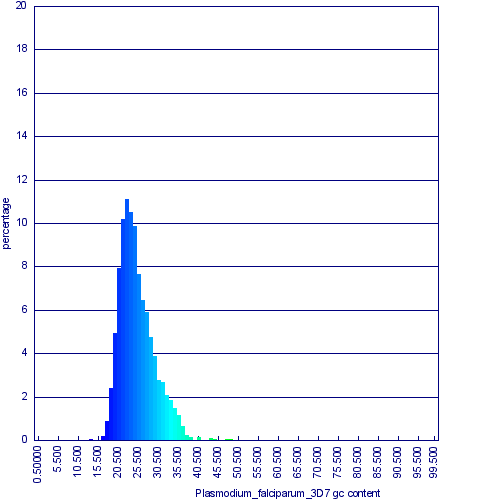


The genomes of two Plasmodium species, falciparum and knowlesi are structurally very similar to one another:
| Organism | Chromosome count | Genome Length | CDS count | Genome GC content | CDS GC content | CDS Wobble position content | non-coding GC content | Plasmodium falicparum | 14 | 22,860,235 bp | 5267 | 19.88% | 23.72% | 17.30% | 14.58% | Plasmodium knowlesi | 14 | 23,462,187 bp | 5102 | 38.94% | 40.23% | 45.56% | 35.12%
} |