Syntenic path assembly
- Syntenic Path Assembly in SynMap of E. coli
-
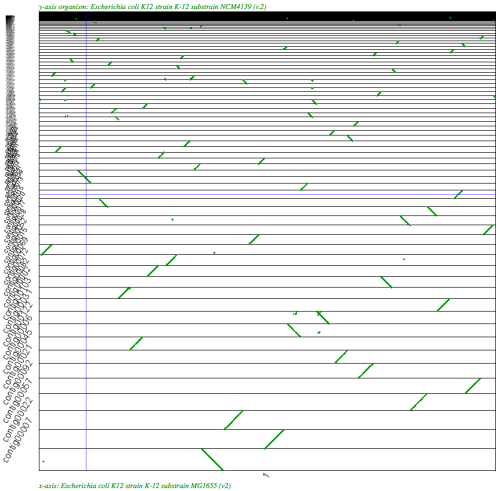
SynMap analysis of a WGS assembly of a strain of E. coli K12 (y-axis) to a reference assembly (x-axis). Results may be regenerated at http://genomevolution.org/r/2k70
-
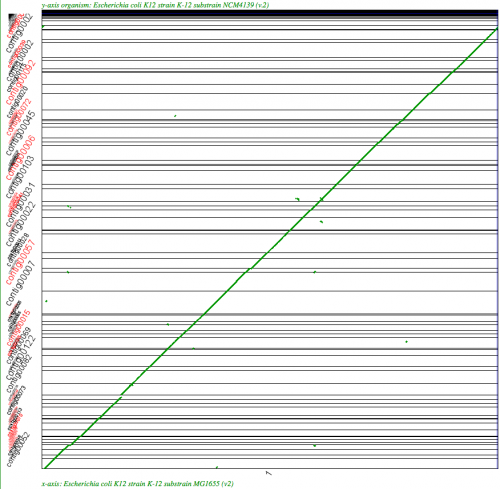
SynMap analysis of a WGS assembly of a strain of E. coli K12 (y-axis) to a reference assembly (x-axis) using SynMap's Syntenic Path Assembly to order contigs. Results may be regenerated at: http://genomevolution.org/r/2k71
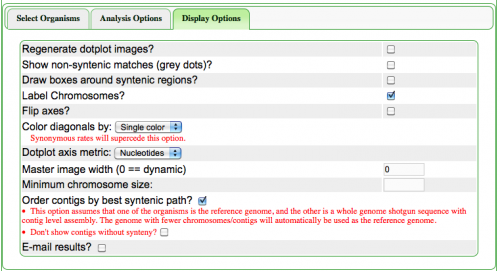
The Syntenic Path Assembly is an option in SynMap to do a reference genome assembly of contigs using synteny to determine the order and orientation of the contigs. To use this option, select "Order contigs by best syntenic path" under the Display Options tab.
-
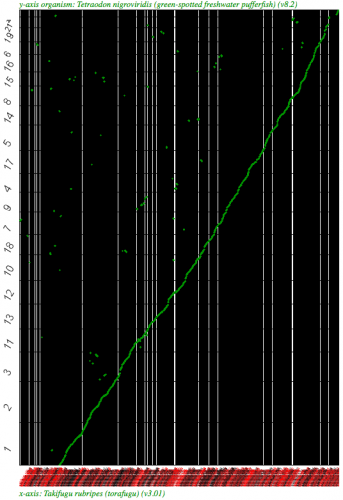
Syntenic dotplot using SynMap between two puffer fish, Takifugu rubripes (x-axis) and Tetraodon nigroviridis (y-axis). Fugu is a WGS assembly. Results may be regenerated at http://genomevolution.org/r/2k7b
-
Syntneic dotplot using SynMap between two puffer fish, Takifugu rubripes (x-axis) and Tetraodon nigroviridis (y-axis) using the Syntenic Path Assembly option. Fugu is a WGS assembly and fully syntenic converage to Tetraodon is detectable. Results may be regenerated at http://genomevolution.org/r/2k79