EPIC-CoGe Tutorial
Overview
This tutorial is aimed at providing an introductory overview of new features in CoGe for the EPIC-CoGe system. These include:
- CoGe User accounts
- Adding private/restricted Experiment data (in this case, epigenetic data)
- Creating Notebooks of collections of data
- Visualizing experimental data with CoGe's implementation of JBrowse
- Sharing data with collaborators
Each section has a brief overview of the process and a video on how to do it.
User accounts

The first thing you will want to do is get a CoGe user account. This is very easy:
- Register an iPlant account
- Log in
More details on how to get a CoGe account
By having a user account, you will be able to:
- Add private data to CoGe
- Share private data with other users
- Create notebooks (collections of data -- experiments and genomes)
- Have CoGe track your analytical history (makes it easy to revisit old analyses)
Adding private data




Once you have a CoGe account, you can add new data to CoGe. To do this:
- Go to your User profile (click on "My Profile" located in the menu at the top-right of any CoGe page)
- Click on the "Create" button to get a menu of things you can create
- Select:
- Load Genome: to load in fasta sequences for a genome
- Load Annotation: to add genome annotations in GFF to a previously loaded genome
- Load Experiment: to add quantitative data to an experiment
Example with an Arabidopsis Ago4 mutant
Here we are going to load in an experiment that is based on bisulfite sequencing of Arabidopsis mutants generated by Steven Jacobsen (ref). You can download the data used for this example here.
- Go to your user profile page: (quick link: http://genomevolution.org/CoGe/User.pl)
- Click "Create" located on the left of the page
- Select "Load Experiment". This will link you to LoadExperiment. (quick link: http://genomevolution.org/CoGe/LoadExperiment.pl)
- When LoadExperiment loads, you will have to enter information about the experiment
- Name: CHH_ago4_Stroud_2013
- Description: CHH methylation
- Version: 1
- Source: Jacobsen Lab
- Note: First search to see if a data source exists. If it does not, you can create a new one by pressing "new".
- Restricted: Check!
- Genome: Arabidopsis thaliana Col-0 (thale cress) (v10.01, id 16911): unmasked
- An experiment must be linked to a genome. Type in part of the name of an organism to start searching for matching genomes.
- Next comes the part of selecting a data file for the experiment. There are three ways to submit your data file to CoGe:
- Upload from your computer
- Fetch from the IPlant Data Store
- Fetch from a remote http or ftp server
- After the experiment has been added, you can manage its information in ExperimentView:
- Change its name, description, source and version
- Add experiment types to it (e.g. BS-Seq)
- Make it public or private
- Add annotations: these are additional descriptors (text, images, and links) for the experiment
- The experiment now shows up in your User profile
- Note: If the experiment was marked as being restricted, a "®" shows up next to its name
Creating Notebooks to manage collections of data



CoGe's Notebooks are designed to make it easy to organize a set of data (genomes, Experiments, Genomic features) and add additional information/Metadata in the form of annotations to the collection of data. In addition, notebooks can be shared with other users of CoGe and are also used to bundle data together in various tools. For example, the EPIC-CoGe Browser uses notebooks to display multiple Experiments in the same track.
To create a notebook
- Go to your user profile page. (quick link: http://genomevolution.org/CoGe/User.pl)
- Click the "Create" button located on the right side of the page and select "New Notebook".
- A box will appear titled "Create New Notebook". Here, type in a name, description (optional), and select the type.
- Type is to help you organize your data.
- Click the red "Create Notebook" to create the notebook.
- The notebook will now in the list of your items. You can search for it by name using the text box located at the top right of the list, or only show notebooks by click on the notebook icon.
- If you hover over the Notebook, information about the notebook appears to the right of the "my stuff" list.
- To add information to the notebook, click on the notebook which will take you to NotebookView. (quick link: http://genomevolution.org/CoGe/NotebookView.pl?nid=503)
- In NotebookView, click "Add Items" to add items:
- When the "Add Items to Notebook" popup box appears, you can select the types of data to search and then use the textbox to type in a search tearm.
- The different tabs limit which sets of data to search -- my stuff are things you have added or have been given access, the others are for public data
- Find the experiment you previously added to CoGe. In this example, the experiment was named "ago4_mutant". Once you find it, select it and press the "Add Selected Items" button.
- Next, search for additional "Ago" mutants by search through the public experiments and add them to your notebook.
- Next, add an annotation to the notebook by pressing the "Add Annotation" button and filling out the popup box.
- In the example shown, the annotation included a figure from a Plant Physiology article on gene silencing movement in Arabidopsis.
To visualize experiments in the [EPIC-CoGe Browser]]
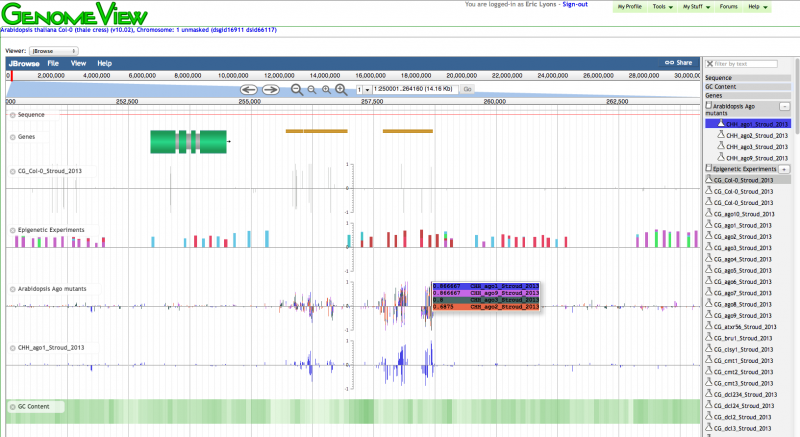
To visualize genomic data in CoGe, you need to go to CoGe's genome browser. Part of the EPIC-CoGe project was to develop a a genome browser that would make it easy for people to overlay Quantitative data on a genome. One challenge to this is how to design an interface that scales well to hundres if not thousands of experiments. To do this, the CoGe team has integrated JBrowse. Besides from the wealth of features JBrowse brings, the CoGe team has developed extensions that mimic the look of the original genome browser in CoGe as well as an hooks to CoGe's experiments, Notebooks, and User-data management system.
- To launch CoGe's genome browser, click on an experiment to launch ExperimentView. (quick link: http://genomevolution.org/CoGe/ExperimentView.pl?eid=330)
- From ExperimentView, click on the link to OrganismView for the genome to which the Experiment is associated. This link is located next to the "Genome:" in the "Experiment info" box. For the link listed above, the genome is "Arabidopsis thaliana Col-0 (thale cress) (v10.02, id16911): unmasked". (quick link: http://genomevolution.org/CoGe/OrganismView.pl?gid=16911)
- Scroll to the bottom of OrganismView and launch the genome browser by clicking the button "Launch Genome Viewer". (quick link: http://genomevolution.org/CoGe/GenomeView.pl?z=6&x=20000&gid=16911&chr=1)
- Currently, when GenomeView loads, it will load the CoGe's original genome browser. To switch to the JBrowse version, select "JBrowse" from the menu next to "Viewer" located above the genome view port. (quick link: http://genomevolution.org/CoGe/GenomeView.pl?z=6&x=20000&gid=16911&chr=1;viewer=JBrowse)
- Note: the quick link will automatically load the JBrowse viewer
- Note: this behavior may change in the future to launch the JBrowse version of the genome browser.
- Information on how to use JBrowse may be found: http://jbrowse.org/
- CoGe specific changes to JBrowse include:
- CoGe-style gene models
- CoGe-style GC content track
- Tracks selection located to the right
- Hooks to CoGe's experiments to visualize experiments and Quantitative data
- Hooks to CoGe's notebooks to visualization multiple sets of experiments in the same track
- Ability to dynamically transform tracks showing multiple experiments (e.g. calculate average values)
Video demonstration of EPIC-CoGe browser
Sharing data in CoGe
Sharing data in CoGe is very easy. There are two general ways to do it:
- Share data directly with another user
- Share data with a User group group of users
To share:
- Go to the user profile page by clicking "My Profile" from the menu located on the top-right of every CoGe page. (quick link: http://genomevolution.org/CoGe/User.pl)
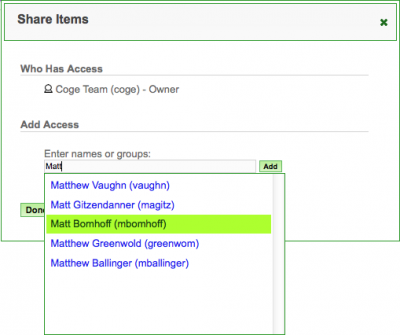
- Select the check-box next to data item(s) your wish to share from the list under "My Stuff"
- Click on the share button
 at the top of the list
at the top of the list - Search and select the person or User-group with whom you wish to share (see right).
- Click "Add" to add that person or User-group
- Click "Done" when you are finished.
- If the person you are seeking is not there, tell them to Get a CoGe account
Video of process: