Syntenic dotplot
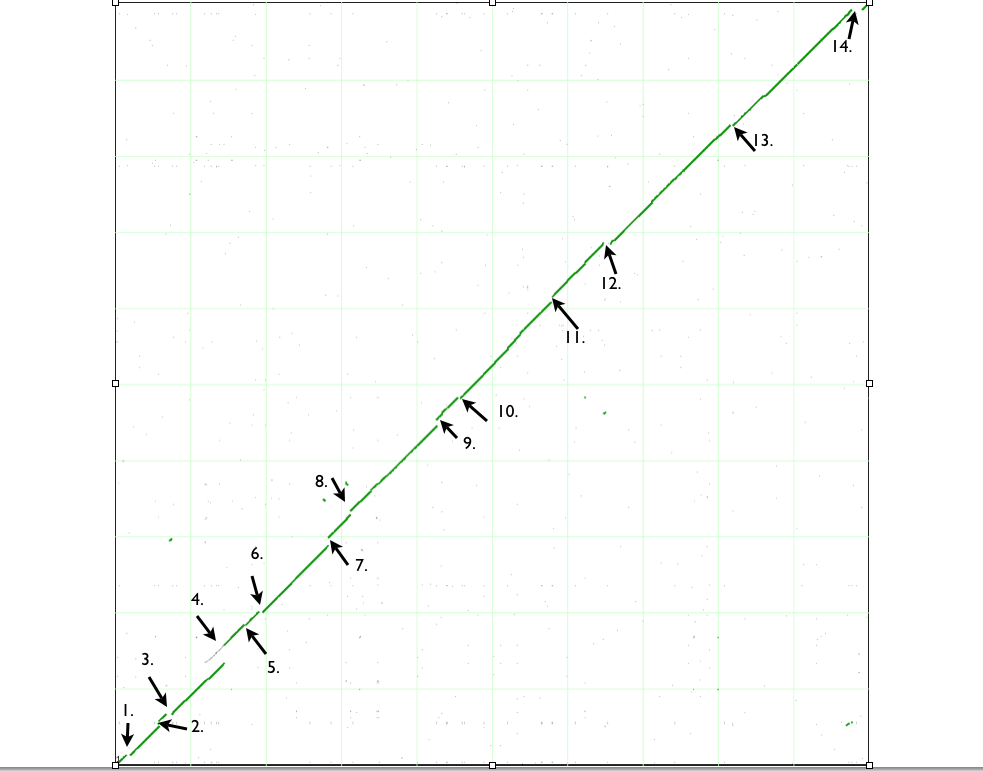
| Variation type |
Difference in strain B REL606 |
Difference in strain K-12 DH10B |
Evidence |
Notes |
Link leading to GEvo |
| 1. Deletion |
none |
Deletion of ~18 genes including DNA pol II, genes in metabolic pathway, thiamine ABC transporter |
pseudogenes in DH10B at deletion site. |
Possible additional insertion in DH10B as evidenced by pseudogenes of yabP, RNA pol associated helicase and FruR, that are not present in REl606 |
tinyurl.com/yexrzpb |
| 2. Insertion |
Insertion of IS1 sequence and a "predicted" transposase |
Insertion sequences and Prophage CP46 DNA insertion | Prophage specific genes found |
Prophage DNA insertion created pseudogenes in K-12 DH10B |
tinyurl.com/yd2quy7 |
| 3. Deletion in B strain or Insertion in K12 strain |
Insertion of IS1 sequence. Insertion of ~15 genes including lac operon and other metabolic enzymes genes | Insertion of IS3 and IS2 sequence | Pseudogene made in DH10B by IS sequence and transposon insertion |
Possible insertion in B REL606 as no evidence of pseudogene is seen in DH10B to account for gene loss in DH10B |
http://tinyurl.com/yedn5tb |
| 4. Insertion in B strain and DNA duplication event in K12. |
Prophage DNA and transposase insertion |
Recent DNA duplication event | 100% identity between paralogs and ~98% identity between syntenic region in REL606 |
Possible phage DNA insertion in REL606 as "hypothetical protein" were found near putative prophage tail component gene in REL606. |
tinyurl.com/yea8bu6 |
| 5. Insertion |
Bacteriophage DNA insertion |
IS2 sequence insertion |
Pseudogenes at IS2 insertion site in DH10B. Phage specific genes were found in REL606 |
Possible page DNA insertion in REL606 as "Hypothetical proteins" were found near phage specific genes |
tinyurl.com/yevlb2w |
| 6. Insertion |
Prophage DNA insertion |
none |
none |
tinyurl.com/ybokuag | |
| 7. Insertion |
none | Prophage DNA insertion | none |
tinyurl.com/yaxlh7o | |
| 8. Insertion of transposon in K12 and B strain. Deletion of phenylacetic acid genes in B strain or insertion of those in K12. |
Rac prophage DNA insertion in K12. Insertion of IS3 element protein and transposase in B strain. Insertion of IS30 transposon in B strain created pseudogene. Presence of phenylacetic acid degradation genes in K12 | tinyurl.com.ye7wcxe | |||
| 9. Insertion in K12 strains. Flagella synthesizing genes either deleted in B strain or inserted in K12. |
Presence of flagella encoding/regulatory genes in K12. IS5 sequence insertion in K12. CP4-44 prophage DNA insertion in K12. | http://tinyurl.com/ybh84x7
| |||
| 10. Insertion in B strain. Pili associated genes and metal resistance genes either inserted in K12 or deleted in B strain. |
Bacteriophage DNA insertion in B strain. IS1 sequence insertion in B strain. Presence of pili associated genes in K12 together with genes conferring resistance to Co and Ni. |
http://tinyurl.com/yammj7v | |||
| 11. Insertion in K12 strain. ParB family protein and recombinase deleted in K12. |
CP4-57 prophage DNA insertion in K12 strain. Presence of ParB family protein gene and recombinase in B strain |
http://tinyurl.com/ycdog2c | |||
| 12. Insertion in B and K12 strain. Inversion. Insertion of saframycin synthetase in B strain or deletion of that in K12. |
Prophage DNA insertion in B strain. Inversion of prepilin leader peptidase/methylase and M-type protein. Is5 insertion in K12. Presence of saframycin synthetase gene in B strain.
|
http://tinyurl.com/ycoagxh | |||
| 13. Insertion of transposon, ShiA-like, TrbC-like proteins in B strain. |
IS30 transposon insertion in B strain. Presence of ShiA-like and TrbC-like protein in B strain |
http://tinyurl.com/ydkrcv8 | |||
| 14. Insertion in B strain |
Insertion of fimbriae synthesis genes, multi drug transporters, Is1 protein gene in B strain. |
http://tinyurl.com/ycwsmsl |