UGTs through the genus Brassica
Introduction
The genus Brassica
The genus Brassica consists of over thirty wild species and hybrids or morphotypes. Generally, species from the genus Brassica are used in food like broccoli, cauliflower, cabbage and more.
The Brassica genome has undergone more polyploidy than Arabidopsis thaliana. Arabidopsis thaliana is notable for being a model organism because of its complexity paired with a relatively small genome.
Duplication Events
The Brassica genome has undergone two tetraploidy and two hexaploidy events, one more than Arabidopsis, since the eudicot paleohexaploidy event which gave rise to Vitis, Prunus, Arabidopsis, and Brassica.
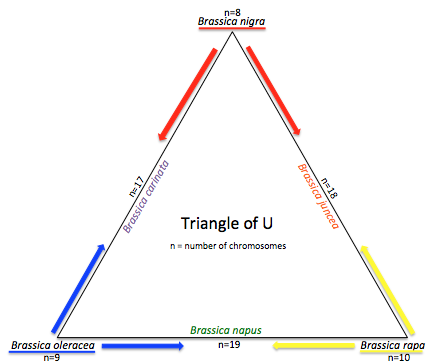
Triangle of U
The "Triangle of U" theory describes the genetic relationship between six species of Brassica: Brassica rapa, Brassica nigra, Brassica oleracea, Brassica juncea, Brassica carinata, and Brassica napus. B. juncea, B. carinata and B. napus are allotetraploids, hybrids with four times the chromosome set of haploids.
UGT Gene Family
UGT functions
Uridine diphosphate (UDP) glycosyltransferases (UGTs) mediate transfer of glycosyl residues from activated nucleotide sugars to acceptor molecules (Tang, Unleashing the Genome of the Brassica rapa). They provide instructions for making enzymes that perform the process of glucuronidation, the addition of glucuronic acid to a substrate (Genetics Home Reference, UGT gene family). This pathway is particularly important in metabolism, and many regard the UGT enzyme as the most important enzyme in the pathway. In humans, these enzymes are responsible for the breakdown of several prescription drugs and pollutants.
UGT chemistry
By mediating transfer of glycosyl residues from activated nucleotide sugars to acceptor molecules, UGTs regulate properties of those acceptors such as bioactivity, solubility and transport within cells and throughout organisms (Ross, Higher plant glycosyltransferases). The UGT enzymes in the metabolic process of glucuronidation, a very common process in Phase II metabolism.
Purpose
UGTs are vital to metabolism of all organisms. Several UGT genes of Arabidopsis thaliana have been sequenced already. Looking into the sequences of Arabidopsis lyrata and Brassica rapa, I hope to determine the exact ratio of UGT genes in each of the species, discover similarities among phylogenetic trees, and pinpoint which genes were conserved, lost, and altered.
Hypotheses
According to how Arabidopsis lyrata and Brassica rapa diverged from Arabidopsis thaliana, and which duplication events occurred in that time, a ratio exists that would explain the UGT genes for each species if there were no losses of genes over time. That ratio looks something like: 1:1:3 of A. thaliana, A. lyrata, and B. rapa respectively. However, looking at the preliminary data we can access from CoGe, Phytozome, and BRAD, those ratios are not what we observe.
Expected Outcome
I expect Arabidopsis lyrata to have approximately the same number of UGT genes as Arabidopsis thaliana with some polymorphisms and possibly some inversions. Brassica rapa which has undergone a further triplication will likely have approximately three times as many UGT genes as A. thaliana with a significantly higher rate of loss than Arabidopsis lyrata. Some polymorphisms and inversions are also expected. In order to determine these ratios, a new phylogenetic tree will be constructed of the UGT genes in Arabidopsis thaliana, Arabidopsis lyrata, and Brassica rapa. Areas of interest within the tree will be further analyzed.
Methods
Building a Phylogeny of Genes in the UGTs in Arabidopsis thaliana
CoGe BLAST
TAIR
Glycosyltransferase Family 1 on The Arabidopsis Information Resource (TAIR) contained each annotation by the The Institute for Genomic Research (TIGR) for flavonols and anthocyanidins which contribute to plant pigmentation. CoGe BLAST was used to find sequences corresponding to those in TAIR. After a little coding, we were able to identify from a list of over a hundred which were from the TAIR database and which were from CoGe with ease. Information including genomic locus, TIGR Annotation and Accession are in appropriately named csv files.
The FASTA sequence for gene At5g65550 was used as a query sequence in the JGI Phytozome database to recover Arabidopsis lyrata genes. The Brassica Database (BRAD) was used to recover orthologs for the Brassica rapa genes.
Organizing the Data
A table was developed to organize the information collected from each database (CoGe, Phytozome, BRAD). 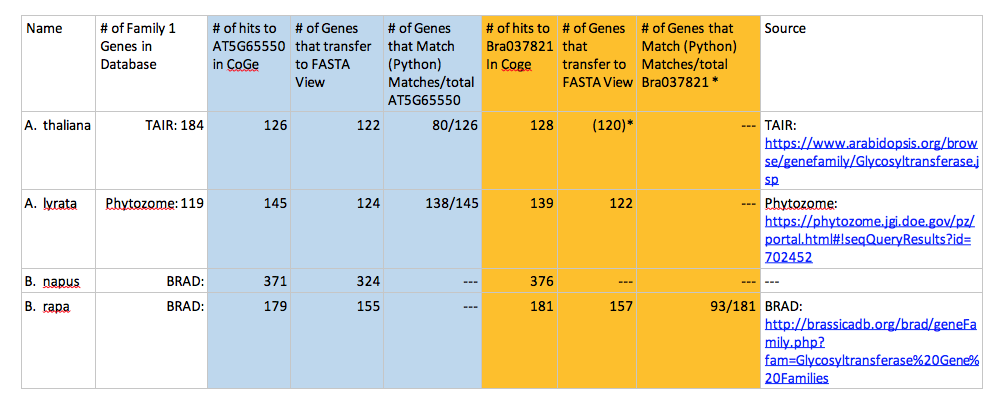
The table above tracks how the size of data changed along the process of collecting the FASTAs. The blue section of the table denotes information relative to the gene At5g65550 while the orange section of the table denotes information relative to the Brassica rapa ortholog to At5g65550, Bra037821.
Determining Test Groups
Of the 28 Glycosyltransferase Families that The Arabidopsis Information Resource (TAIR) has on Arabidopsis thaliana, I chose to work with Family 1 due to the fact that most of the genes were similar in function as the TIGR Annotation suggested. A preliminary tree was constructed of 122 of these genes (those that CoGe had FASTA sequences for). With use of Keiko Yonekura-Sakakibara’s Functional genomics of family 1 glycosyltransferases in Arabidopsis, I began identifying several functions of the genes. We identified three clusters on the tree that we named Test groups. Test group 1 consists of AT2G36750, AT2G36760, AT2G36770, AT2G36780, AT2G36790, and AT2G36800. These genes exhibit functions of flavonol 7-O-glucosyltransferase and brassinosteroid O-glucosyltransferase. Test group 2 consists of AT3G21780, AT4G15720, AT4G15260, AT4G15280, AT3G21750, and AT3G21760. These genes exhibit the function of having ABA glucosyltransferase activity. Test group 3 consists of AT4G01070, AT1G01420, AT1G01390, AT3G50740, AT5G66690, AT5G26310, AT2G18570, AT2G18560, AT4G36670. These genes exhibit the function of monolignol 4-O-glycosyltransferase activity and having xeniobiotic glycosyltransferase activity. Each of these test groups genes’ FASTAs were placed into one file. Using GeVo, each Arabidopsis thaliana gene was visualized for syntenic regions in Arabidopsis lyrata and Brassica rapa. The FASTAs for those genes in were added to the file and the gevolinks were saved to a separate files for each test group. Using phylogeny.fr, new trees were constructed with Arabidopsis thaliana, Arabidopsis lyrata, and Brassica rapa for each test group.