Source code
Software licenses
Individual tools used in CoGe are subject to their own licenses, and are developed independently of CoGe
CoGe's components
CoGe's software is divided into several components:
- Database (mysql)
- Database API (perl)
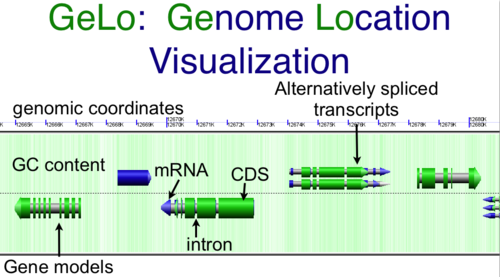
- Genome Location Visualization: GeLo (perl)
- Website and applications (perl, javascript, python, flash, HTML)
- Other stuff: alignment algorithms, etc.
As documentation often lags behind development, much of CoGe's software is not in a suitable condition for release. However, once it has been, we will post its code here. Also, we will be happy to provide any code as requested and do our best to answer any questions you may have about it.
CoGe's genome visualization library GeLo
NWAlign
nwalign is the implementation of the Needleman-Wunsch algorithm for global sequence alignments that CoGe uses. This very fast and easily parallelized implementation is written and maintained by Brent Pedersen and can be downloaded at: http://pypi.python.org/pypi/nwalign/
python tools for DAGChainer
Brent Pedersen has written an extensive library of tools, including a rewrite of DAGChainer's perl front-end, in python. These tools greatly extend the functionality of DAGChainer. For example, an automated way to merge syntenic blocks by recoding output from a DAGChainer run and resubmitting it back to DAGChainer. These tools can be obtained though an svn repository:
svn co http://bpbio.googlecode.com/svn/trunk/scripts/dagchainer