Syntenic dotplot of peach versus grape


Please note: This genome is currently unpublished and therefore under the publication restrictions of the Fort Lauderdale Convention.
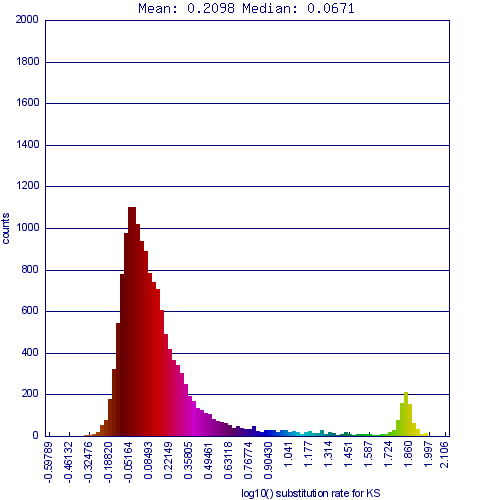
Whole genome syntenic comparison of peach tree's genome to grapevine's shows extensive synteny where each region of each genome matching one other region very well, and two other regions less well. While the colors are a bit difficult to differentiate in this image, the better matching regions are a deeper red, signifying a lower Ks value. These regions are derived from the divergence of these lineages and are orthologs, the other regions being out-paralogous and derived from their shared eudicot paleohexaploidy event.
This analysis also shows that peach, like grapevine, has not undergone a subsequent whole genome duplication event. Overall, the structure of these genomes is very similar as evidenced by the large syntenic orthologous regions. However, peachtree's lower chromosome number is likely due to chromosome fusion events of chomosomes that have mostly remained separated in grapevine.
High-resolution sequence analysis of syntenic regions between these genomes shows extensive conservation of genes and gene order. These analyses help to identify tandem gene duplications, inversions, and true orthologous genes.