Cacao syntenic dotplots


Syntenic dotplot between Cacao and Peach genomes
The syntenic dotplot shows a strong 1:1 mapping of syntenic regions, and weaker 3:3 mappings of syntenic regions. This is due to the shared paleohexaploidy event at the base of the eudicots and the lack of subsequent whole genome duplications events in either lineage.
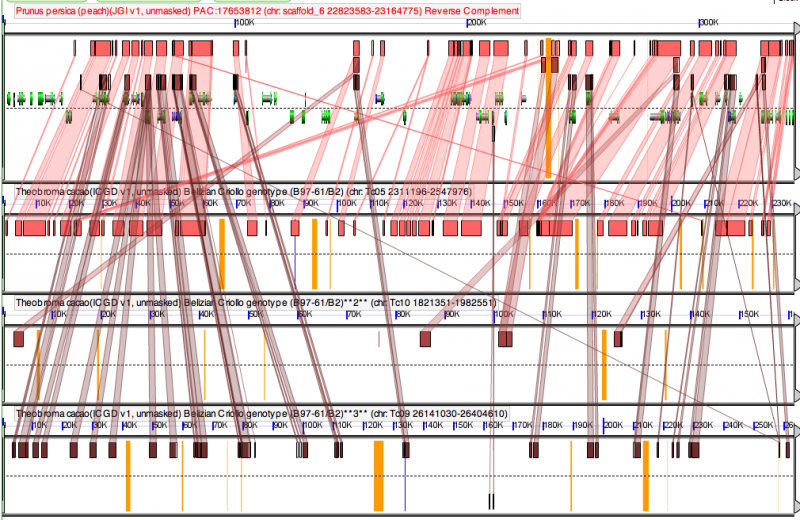
High-resolution syntenic analysis of Cacao and Peach genomes
The GEvo analysis of sytnenic regions between cacao and peach (top pannel) shows that one cacao region is highly syntenic to a peach region. These regions are derived from the divergence of the peach and cacao lineages. The other two syntenic cacao regions show much weaker synteny (fewer collinear homologous regions). These regions are derived from the eurosid paleohexaploidy.