Sequenced plant genomes
This site attempts to track all plant genomes with published sequences, and at least some of the genomes currently in the process of being sequenced. Genomes are divided into four states:
- Published: A complete genome sequence is available, and anyone can publish papers on it without restriction.
- Unpublished: The complete sequence (or a substantially complete sequence) is available, but whole genome analysis cannot be published until the group that sequenced the genome publishes their own paper describing it. These restrictions are outlines by the Fort Lauderdale Convention.
- Incomplete: A partial assembly is available, but sequencing and/or assembly and/or gene annotation is ongoing.
- Unreleased: Genome sequencing has at least begun, but no data has been made publicly available.
Phylogenetic Tree
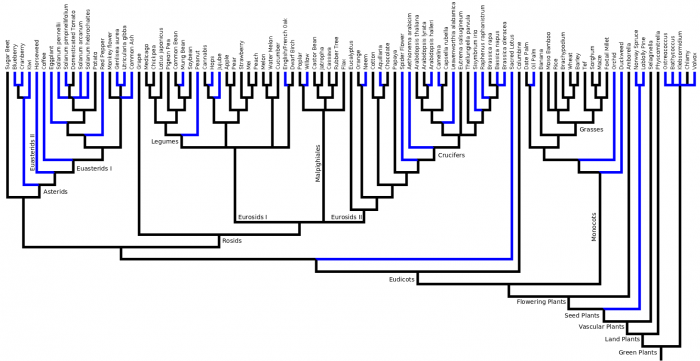
Eudicots
The eudicots are the largest group of flowering plants on the planet.
Columbine
(genome available but unpublished)
Columbine (Aquilegia sp.) comes from a group of eudicots, the Ranunculales, whose ancestors split from the ancestors of the major eudicot groups (like rosids and asterids) a long, long, time ago (somewhere in the neighborhood of 115-130 million years ago). Comparing the columbine genome sequence with other eudicot genomes should be very interesting for several groups of plant biologists (comparative genomicists and evolutionary biologists in particular).
The columbine genome was sequenced to 8-fold coverage by JGI and a pre-publication release of the genome is available for download from phytozome. The current assembly is only to the scaffold level (no pseudomolecules) and consists of 302 megabases of sequence spread over 971 scaffolds. Current gene annotations identify 25,784 genes identified by a mixture of EST sequencing and homology to other sequenced genomes.
Asterids
The asterids are a group of plants within the eudicots that include species like the solanacious vegetables (Tobacco, Tomato, Potato, Peppers and Eggplant) and the sunflowers. There are not currently any published plant genomes from species in the Asterid clade, but there are at least three genome projects with some level of sequence publicly available, and the sunflower genome project was recently announced. (See planned, in-progress, and private genomes at the bottom of this page)
Tomato
(genome incomplete)
The tomato (Solanum lycopersicum) genome project is not yet complete. The version of the genome currently loaded into CoGe is assembled into pseudomolecules[1] but does not contain [2]. The most recent assembly is 1.03 which is assembled from 22x coverage sequencing using 454 technology. Read more about the tomato genome project here or see it in GenomeView here.
Potato
(genome incomplete)
The potato genome project is not yet complete. Read more about the potato genome projector see it in GenomeView here.
Monkey Flower
(genome incomplete)
The monkey flower (Mimulus guttatus) genome is not yet complete. The version of the genome currently loaded into CoGe is not assembled into pseudomolecules[1] but does contain genome models[2] Read more about the monkey flower genome on phytozomeor see the current assembly in GenomeView here.
Rosids
Grape

The genome sequence of the wine grape (Vitis vinifera) was published by a group of French and Italian researchers in 2007. The variety of grape sequenced was the Pinot Noir.
Grape diverged early from the two main groups of species in the rosids (eurosids I and eurosids II) and has not experienced any whole genome duplications since that divergence making it an important outgroup for comparisons to other rosid species as well as providing a great resource for studying the ancient hexaploidy that preceeding the radiation of rosid species (and possibly the radiation of eudicot species).
The version of the grape genome in CoGe contains ~500 megabases of sequence and 26346 annotated genes spread across 19 chromosomes.
The genome paper:
Jaillon, O., Aury, J., Noel, B., Policriti, A., Clepet, C., Casagrande, A., Choisne, N., Aubourg, S., Vitulo, N., Jubin, C., Vezzi, A., Legeai, F., Hugueney, P., Dasilva, C., Horner, D., Mica, E., Jublot, D., Poulain, J., Bruyère, C., Billault, A., Segurens, B., Gouyvenoux, M., Ugarte, E., Cattonaro, F., Anthouard, V., Vico, V., Del Fabbro, C., Alaux, M., Di Gaspero, G., Dumas, V., Felice, N., Paillard, S., Juman, I., Moroldo, M., Scalabrin, S., Canaguier, A., Le Clainche, I., Malacrida, G., Durand, E., Pesole, G., Laucou, V., Chatelet, P., Merdinoglu, D., Delledonne, M., Pezzotti, M., Lecharny, A., Scarpelli, C., Artiguenave, F., Pè, M., Valle, G., Morgante, M., Caboche, M., Adam-Blondon, A., Weissenbach, J., Quétier, F., & Wincker, P. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla Nature, 449 (7161), 463-467 DOI: 10.1038/nature06148
Rose Gum Tree
(genome unpublished)
One of several species of tree referred to by the common name "Eucalyptus", the rose gum tree (Eucalyptus grandis) is native to Australia, but is considered a candidate for biofuel production in the US. The rose gum tree is a basal rosid, like grape, so in addition to the value of this genome sequence for biofuel breeding purposes, this genome serves as a valuable outgroup for the core rosids (listed as Eurosids 1 and Eurosids 2 on this site).
The rose gum genome was sequenced to 8x coverage by the Joint Genome Institute and is assembled into 11 linkage/chromosome groups. The initial release of the genome includes 691 MB of sequence data, and 41,204 protein coding genes located on the putative chromosome assemblies. Read more/download the sequence from Phytozome.
Eurosids 1
Cucumber

The genome sequence of cucumber (Cucumis sativus) was published in late 2009. The genome of the inbred line "'Chinese long' 9930" was sequenced using a combination of Illumina short read sequencing (68.3x coverage) and Sanger sequencing (3.9x coverage). The complete published genome consists of 243.5 megabases of sequence and 26,682 protein coding genes, 72.8% of which can been anchered to the seven cucumber chromosomes, with the rest unanchored.
The publication also included a genetic map with a total length of 581 cM based on 1,885 markers. Resources from this version of the cucumber genome are available at this site.
Independently a group of researchers in the US have released a draft of a cucumber genome sequence of the inbred line Gy14. This version of the genome was assembled from 454 sequencing reads and the current release consists of 203 megabases of sequence and a predicted 21491 protein coding genes spread over 4219 scaffolds. This version is available at Phytozome.
The genome paper: Huang, S., Li, R., Zhang, Z., Li, L., Gu, X., Fan, W., Lucas, W., Wang, X., Xie, B., Ni, P., Ren, Y., Zhu, H., Li, J., Lin, K., Jin, W., Fei, Z., Li, G., Staub, J., Kilian, A., van der Vossen, E., Wu, Y., Guo, J., He, J., Jia, Z., Ren, Y., Tian, G., Lu, Y., Ruan, J., Qian, W., Wang, M., Huang, Q., Li, B., Xuan, Z., Cao, J., Asan, ., Wu, Z., Zhang, J., Cai, Q., Bai, Y., Zhao, B., Han, Y., Li, Y., Li, X., Wang, S., Shi, Q., Liu, S., Cho, W., Kim, J., Xu, Y., Heller-Uszynska, K., Miao, H., Cheng, Z., Zhang, S., Wu, J., Yang, Y., Kang, H., Li, M., Liang, H., Ren, X., Shi, Z., Wen, M., Jian, M., Yang, H., Zhang, G., Yang, Z., Chen, R., Liu, S., Li, J., Ma, L., Liu, H., Zhou, Y., Zhao, J., Fang, X., Li, G., Fang, L., Li, Y., Liu, D., Zheng, H., Zhang, Y., Qin, N., Li, Z., Yang, G., Yang, S., Bolund, L., Kristiansen, K., Zheng, H., Li, S., Zhang, X., Yang, H., Wang, J., Sun, R., Zhang, B., Jiang, S., Wang, J., Du, Y., & Li, S. (2009). The genome of the cucumber, Cucumis sativus L. Nature Genetics, 41(12), 1275-1281 DOI: 10.1038/ng.475
Poplar

The genome sequence of the black cottonwood tree (Populus trichocarpa) was published in 2006. The genome was originally sequenced to a coverage of 7.5x using Sanger sequencing. Poplar was the third plant genome to be published, and is now one of two published genomes of tree species (the other being papaya). Poplar contains a whole genome duplication that is not shared by any other plant species with a sequenced genome. The most recent version of the poplar genome in CoGe is v2 available on Phytozome which includes ~370 megabases of sequence and 41377 protein coding genes spread over 19 chromosomes.
The genome paper: Tuskan, G., DiFazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., Putnam, N., Ralph, S., Rombauts, S., Salamov, A., Schein, J., Sterck, L., Aerts, A., Bhalerao, R., Bhalerao, R., Blaudez, D., Boerjan, W., Brun, A., Brunner, A., Busov, V., Campbell, M., Carlson, J., Chalot, M., Chapman, J., Chen, G., Cooper, D., Coutinho, P., Couturier, J., Covert, S., Cronk, Q., Cunningham, R., Davis, J., Degroeve, S., Dejardin, A., dePamphilis, C., Detter, J., Dirks, B., Dubchak, I., Duplessis, S., Ehlting, J., Ellis, B., Gendler, K., Goodstein, D., Gribskov, M., Grimwood, J., Groover, A., Gunter, L., Hamberger, B., Heinze, B., Helariutta, Y., Henrissat, B., Holligan, D., Holt, R., Huang, W., Islam-Faridi, N., Jones, S., Jones-Rhoades, M., Jorgensen, R., Joshi, C., Kangasjarvi, J., Karlsson, J., Kelleher, C., Kirkpatrick, R., Kirst, M., Kohler, A., Kalluri, U., Larimer, F., Leebens-Mack, J., Leple, J., Locascio, P., Lou, Y., Lucas, S., Martin, F., Montanini, B., Napoli, C., Nelson, D., Nelson, C., Nieminen, K., Nilsson, O., Pereda, V., Peter, G., Philippe, R., Pilate, G., Poliakov, A., Razumovskaya, J., Richardson, P., Rinaldi, C., Ritland, K., Rouze, P., Ryaboy, D., Schmutz, J., Schrader, J., Segerman, B., Shin, H., Siddiqui, A., Sterky, F., Terry, A., Tsai, C., Uberbacher, E., Unneberg, P., Vahala, J., Wall, K., Wessler, S., Yang, G., Yin, T., Douglas, C., Marra, M., Sandberg, G., Van de Peer, Y., & Rokhsar, D. (2006). The Genome of Black Cottonwood, Populus trichocarpa (Torr. & Gray) Science, 313 (5793), 1596-1604 DOI: 10.1126/science.1128691
Woodland Strawberry
The woodland strawberry (Fragaria vesca) is not the species that produces most of the strawberries you see on grocery store shelves today. Those are generally from the garden strawberry. However garden strawberries are octoploid, making sequencing their genome relatively difficult, while the woodland strawberry possesses a much more manageable diploid genome. For more on the story of how the woodland strawberry came to be sequenced, check out this fascinating story from one of the scientists behind the genome paper.
The published strawberry genome consists of 7 chromosomes/pseudomolecules.
The Genome Paper
Vladimir Shulaev et al., "The genome of woodland strawberry (Fragaria vesca)," Nat Genet 43, no. 2 (February 2011): 109-116.
Castor Bean
The castor bean (Ricinus communis) is an oilseed plant that is the source of castor oil and the deadly poison ricin. The castor bean should not to be confused with the common bean (Phaseolus vulgaris) which is in the Joint Genome Institute sequencing pipeline.
The published castor bean genome is based on a 4.6x coverage of the genome using solexa sequencing.
The current release consists of 31,237 gene models spread across 25,800 scaffolds.
The entire genome is estimated to be ~320 megabases in size and contains 10 chromosomes.
Genome Paper
Agnes P Chan et al., “Draft genome sequence of the oilseed species Ricinus communis,” Nat Biotech advance online publication (online 2010), http://dx.doi.org/10.1038/nbt.1674
The website of the castor bean sequencing group.
Cassava
(genome incomplete)
Cassava (Manihot esculenta) is the most important crop that most people in America and Europe have never heard of (except perhaps in the form of tapioca). Originally domesticated in South America, cassava is now an important food source in Southeast Asia and Africa. The current draft genome is made available through phytozome and consists of 416 megabases of sequence spread over 11,243 contigs. This is only a little over 50% of the estimated total size of the cassava genome, but the people involved in the sequencing and assembly believe it represents the majority of the non-repetitive genome. The current release also includes 47,164 predicted genes.
Apple
The Apple (Malus x domestica) genome was published in late August. The total genome is estimated to be 742.3 MB large, spread over 17 chromosomes. The published genome includes 600 megabases of sequence assembled into 17 pseudomolecules and a number of smaller unanchored contigs. The apple genome contains 57,386 putative genes, a high number attributable, at least in part, to a whole genome duplication in the apple lineage which is dated to 30-65 million years ago. The apple genome is not yet loaded into CoGe, and does not yet appear to be available for download, however, there is an available genome browser.
Genome Paper Riccardo Velasco et al., “The genome of the domesticated apple (Malus [times] domestica Borkh.),” Nat Genet advance online publication (online 2010), [1]
Peach
(genome unpublished)

Peaches (Prunus persica) are stone fruits, meaning they're closely related to fruits such as plums, apricots, and cherries and nuts like almonds. The 1.0 version of the peach genome assembly was released by the International Peach Genome Initiative on April 1st, 2010. This version of the genome is already assembled into eight pseudomolecules covering the eight chromosomes of peach, as well as ~200 smaller unplaced contigs. The total released sequence is 227 megabases and includes 27,852 annotated genes. The genome was sequenced to 7.7x coverage using Sanger sequencing.
Legumes
Legumes (the plant family Fabaceae) contained within the eurosid II clade. The family is perhaps best known for the fact that many of the species it contains form symbiotic relationships with nitrogen fixing bacteria. The bacteria are sheltered and feed within special nodules in the roots of these plants and in return the plant benefits from the bacteria's ability to convert the nitrogen in our atmosphere into bio-available forms (bioavailable nitrogen is often a limiting nutrient for other plant species).
Medicago
(genome unpublished)
Medicago (Medicago truncatula) is small legume used as a model species for nodule formation and nitrogen fixing. The latest release of the medicago genome is Mt3.0 which includes 240 megabases of sequence associated with Medicago's eight chromosomes, plus 16.6 megabases of unanchored sequence. Read more at International Medicago Genome Annotation Group's webpage.
Soybean

Soybeans (Glycine max) are an important crop species valued both as a source of protein and for their ability to fix nitrogen, which reduces the amount of fertilizer that needs to be applied to whatever crop is grown in the same field the following year.
The soybean genome was published in early 2010 and contained 950 megabases of sequence as well as a predicted 46,430 protein coding genes distributed over twenty chromosomes. The ancestors of soybean went through two whole genome duplications since the ancient hexaploidy as the base of the eudicot lineage with the older estimated to have occured 59 million years ago and the more recent estimated to have occured 13 million years ago.
The Genome Paper: Schmutz, J., Cannon, S., Schlueter, J., Ma, J., Mitros, T., Nelson, W., Hyten, D., Song, Q., Thelen, J., Cheng, J., Xu, D., Hellsten, U., May, G., Yu, Y., Sakurai, T., Umezawa, T., Bhattacharyya, M., Sandhu, D., Valliyodan, B., Lindquist, E., Peto, M., Grant, D., Shu, S., Goodstein, D., Barry, K., Futrell-Griggs, M., Abernathy, B., Du, J., Tian, Z., Zhu, L., Gill, N., Joshi, T., Libault, M., Sethuraman, A., Zhang, X., Shinozaki, K., Nguyen, H., Wing, R., Cregan, P., Specht, J., Grimwood, J., Rokhsar, D., Stacey, G., Shoemaker, R., & Jackson, S. (2010). Genome sequence of the palaeopolyploid soybean Nature, 463 (7278), 178-183 DOI: 10.1038/nature08670
Eurosids 2
Chocolate
The genome of the tree that gives us chocolate Theobroma cacao has been independently sequenced by two groups. One genome assembly, of the variety called Criollo from Belize has been published in the journal Nature Genetics (see citation below). A second assembly of a breed called Matina 1-6 has available from the Cacao genome database since before the publication of the Criollo genome sequence, but has not yet been published. Both assemblies are complete to the level of pseudomolecules.
Chocolate has not experienced any whole genome duplications since the ancient hexaploidy shared by all sequenced rosids.
Genome Paper (Criollo version: Xavier Argout et al., "The genome of Theobroma cacao," Nat Genet 43, no. 2 (February 2011): 101-108.
Citrus fruits (genus Citrus)
Citrus fruits from lemons to pomelos belong to a singe genus. Many fruits we think of as separate species can breed with each other, making it difficult to properly define species barriers.
Sweet/Common Orange
(genome unpublished and not fully assembled)
The sweet orange (Citrus sinensis) was sequenced using a combination of Sanger (old fashion, expensive, but long and easy to assemble) and 454 (much cheaper, faster, and somewhat shorter) sequencing technology. The current release is only version 0.1 and the genome is still split into 12,574 scaffolds that cover a combined 319 megabases of the sweet orange genome. Unlike the clementine genome described below, the sweet orange genome project used DNA from a diploid individual, making the assembly of the genome somewhat more difficult as inconsistences between aligned sequences might simply be the result of variation between the two genome copies of that diploid individual. This version of the genome release includes 25,376 annotated protein coding genes. You can read more or download data here.
Clementine mandarin
(genome unpublished and not fully assembled)
The genome of a haploid Clementine orange (Citrus clementina) was sequenced by the International Citrus Genome Consortium to a coverage of 6.5-fold. The genome is not yet assembled into pseudomolecules but consists of 1,128 scaffolds containing a total of 296 megabases of sequence data. Genes were predicted using both sequencing of ESTs and homology to the genes of other sequenced plant species, resulting in a total of 25,385 protein coding genes. Download clementine sequence data and annotations from phytozome here.
Papaya

The genome of the papaya tree (Carica papaya) was published in the early 2008. Papaya was one of the earliest crops to be genetically modified (in papaya's case to resist the devastating papaya ringspot virus) and the sequenced genome actually comes from one of the genetically modified varieties (SunUp). The papaya genome was sequenced to a coverage of 3x using Sanger sequencing. Papaya has not experienced further [whole genome duplications] since the ancient hexaploidy shared by all currently sequenced eudicots. As the most closely related species to Arabidopsis with a currently sequenced genome that has not experienced the two subsequence whole genome duplications found in the Arabidopsis lineage, papaya is a useful outgroup, although the ancestors of Arabidopsis and Papaya split ~72 million years ago.
The papaya genome is estimated to be have a size of 372 megabases, spread across nine chromosomes, and contain (X) genes. The version of papaya within CoGe is organized into super contigs, but does contain a number of gaps.
The genome paper:
Ming, R., Hou, S., Feng, Y., Yu, Q., Dionne-Laporte, A., Saw, J., Senin, P., Wang, W., Ly, B., Lewis, K., Salzberg, S., Feng, L., Jones, M., Skelton, R., Murray, J., Chen, C., Qian, W., Shen, J., Du, P., Eustice, M., Tong, E., Tang, H., Lyons, E., Paull, R., Michael, T., Wall, K., Rice, D., Albert, H., Wang, M., Zhu, Y., Schatz, M., Nagarajan, N., Acob, R., Guan, P., Blas, A., Wai, C., Ackerman, C., Ren, Y., Liu, C., Wang, J., Wang, J., Na, J., Shakirov, E., Haas, B., Thimmapuram, J., Nelson, D., Wang, X., Bowers, J., Gschwend, A., Delcher, A., Singh, R., Suzuki, J., Tripathi, S., Neupane, K., Wei, H., Irikura, B., Paidi, M., Jiang, N., Zhang, W., Presting, G., Windsor, A., Navajas-Pérez, R., Torres, M., Feltus, F., Porter, B., Li, Y., Burroughs, A., Luo, M., Liu, L., Christopher, D., Mount, S., Moore, P., Sugimura, T., Jiang, J., Schuler, M., Friedman, V., Mitchell-Olds, T., Shippen, D., dePamphilis, C., Palmer, J., Freeling, M., Paterson, A., Gonsalves, D., Wang, L., & Alam, M. (2008). The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature, 452 (7190), 991-996 DOI: 10.1038/nature06856
Arabidopsis species and allies
Expect this category to grow substantially over the next year. The planned, in progress, and private genomes category below includes 7 more arabidopsis species and relatives.
Arabidopsis thaliana

Arabidopsis thaliana is a poplar model plant species, partially as a result of its short generation time and compact size. The genome of Arabidopsis was also the first plant genome to be published back in 2000. The current release of the Arabidopsis genome is TAIR10:
The TAIR10 release contains 27,416 protein coding genes, 4827 pseudogenes or transposable element genes and 1359 ncRNAs (33,602 genes in all, 41,671 gene models). A total of 126 new loci and 2099 new gene models were added.
The Arabidopsis genome is ~120 megabases of sequence spread across five chromosomes.
Genome resources:
The 1001 genomes project[3] plans to sequence the genomes of 1001 different varieties of Arabidopsis. Currently 88 are available with more in progress.
The Genome Paper: The Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408 (6814), 796-815 DOI: 10.1038/35048692
Arabidopsis lyrata
(genome unpublished)
Arabidopsis lyrata also has a sequenced, though unpublished, genome. As a close relative of A. thaliana, the lyrata genome is valuable for comparative genomics. A. lyrata also is self-incompatable, while A. thaliana reproduced primarily through self-fertilization. The lyrata genome is available within CoGe, or you can download it from JGI.
Monocots
Date Palm
(genome incomplete)
The date palm genome assembly is VERY incomplete. The sequence was generated by whole genome shotgun sequencing, which included some assembly to join contigs into scaffolds. The current assembly (v2) is available from the date palm website of Weill Cornell Medical College.
Banana
(genome unreleased)
Baring a contender coming out of left field, the banana genome is on track to be the first published non-grass monocot genome. It will be incredibly valuable from a comparative genomics standpoint for studying the evolution of grass genomes. However banana isn't being sequenced to make the lives of comparative genomicists easier, but because of the key role many banana species play in the tropical food production. Bananas are also a target of genetic engineering, since the fact that most cultivated breeds are triploid and unable to reproduce sexually (the reason bananas aren't full of seeds) makes conventional breeding impossible, and bananas suffer from a number of nasty plant pathogens.
As of the 18th Plant and Animal Genome Conference (January 2010) the banana genome project was planning to wrap up sequencing in early 2010, and spend the spring doing gene annotation. The variety of banana they chose for sequencing was "Pahang DH" a breed of Musa acuminata malaccensis with an estimated genome size of 600-700 megabases.
Trivia: The banana is the most consumed fruit in America, with the average American eating ~25 pounds of bananas per year, a full quarter of the average total fruit consumed per person. Yet almost NO bananas are produced domestically.
Grasses
The grasses, a family of plants known as the poaceae, can trace their lineages back to a common ancestor that probably lived between 50-70 million years ago, either right before or soon after the extinction of the dinosaurs(dinosaurs didn't eat grass). Since their emergence in the fossil record, the grasses have been extraordinarily successful, becoming one of the largest families of plants on the planet and covering vast swaths of the planet in the form of prairies/savannahs/steppes.
While you may think of grass primarily as the green stuff on lawns and sports fields, remember that grasses also include species like bamboo and the grains that make up so much of what we eat. Either three (rice, wheat, and corn/maize) or four (the same three plus sugar cane) grass species provide more than half of all the calories that feed the worlds population[4], and are the focus of much applied and basic scientific research. Check out the Pan-grass synteny project
Rice

Rice (Oryza sativa) was the second plant genome (after Arabidopsis) to be published, making it the first monocot genome, the first grass genome, the first food crop genome, and the first grain genome (and probably a whole lot of other firsts as well). The original published genome (published in 2002) was from the subspecies Oryza sativa japonica, however the genome of the other primary subspecies Oryza sativa indica has also been sequenced.
The current version of the rice genome in CoGe is v6.1 from MSU which contains ~370 megabases of sequence and 40,577 non-transposon related genes spread across 12 chromosomes.
Rice Resources:
- The MSU Rice Gene Annotation Project the source of the gene annotations used by CoGe
- RAP-DB: The Rice Annotation Project Database. A different set of annotations for the rice genome
The genome paper:
Goff, S. et al. (2002). A Draft Sequence of the Rice Genome (Oryza sativa L. ssp. japonica) Science, 296 (5565), 92-100 DOI: 10.1126/science.1068275
Brachypodium

The brachy genome (Brachypodium distachyon) was published in early 2010. Brachypodium is a small temperate grass native found around the Mediterranean, and east into India. Its choice as a model organism was based on small physical size, quick generation time, and small genome (a lot of the same reasons as Arabidopsis) as well as its membership in the Pooideae, a group of grass species that also includes important crop species: wheat, barley, rye, and oats all species whose genomes have not yet been sequenced (although the last common ancestor of brachy and these important crop species is estimated to have lived >30 million years ago). Brachy's genome is currently the only published genome of a non-domesticated grass and the only temperate (as opposed to tropical) grass.
The published version of the brachy genome includes 272 megabases of sequence and 25,532 protein coding genes spread across five chromosomes. It was sequenced to a coverage of 9.4× using Sanger sequencing.
Brachy Resources:
- Brachypodium.org is probably the first place to check.
- Brachy's page at JGI
The genome paper:
Vogel, J., Garvin, D., Mockler, T., Schmutz, J., Rokhsar, D., Bevan, M., Barry, K., Lucas, S., Harmon-Smith, M., Lail, K., Tice, H., Schmutz (Leader), J., Grimwood, J., McKenzie, N., Bevan, M., Huo, N., Gu, Y., Lazo, G., Anderson, O., Vogel (Leader), J., You, F., Luo, M., Dvorak, J., Wright, J., Febrer, M., Bevan, M., Idziak, D., Hasterok, R., Garvin, D., Lindquist, E., Wang, M., Fox, S., Priest, H., Filichkin, S., Givan, S., Bryant, D., Chang, J., Mockler (Leader), T., Wu, H., Wu, W., Hsia, A., Schnable, P., Kalyanaraman, A., Barbazuk, B., Michael, T., Hazen, S., Bragg, J., Laudencia-Chingcuanco, D., Vogel, J., Garvin, D., Weng, Y., McKenzie, N., Bevan, M., Haberer, G., Spannagl, M., Mayer (Leader), K., Rattei, T., Mitros, T., Rokhsar, D., Lee, S., Rose, J., Mueller, L., York, T., Wicker (Leader), T., Buchmann, J., Tanskanen, J., Schulman (Leader), A., Gundlach, H., Wright, J., Bevan, M., Costa de Oliveira, A., da C. Maia, L., Belknap, W., Gu, Y., Jiang, N., Lai, J., Zhu, L., Ma, J., Sun, C., Pritham, E., Salse (Leader), J., Murat, F., Abrouk, M., Haberer, G., Spannagl, M., Mayer, K., Bruggmann, R., Messing, J., You, F., Luo, M., Dvorak, J., Fahlgren, N., Fox, S., Sullivan, C., Mockler, T., Carrington, J., Chapman, E., May, G., Zhai, J., Ganssmann, M., Guna Ranjan Gurazada, S., German, M., Meyers, B., Green (Leader), P., Bragg, J., Tyler, L., Wu, J., Gu, Y., Lazo, G., Laudencia-Chingcuanco, D., Thomson, J., Vogel (Leader), J., Hazen, S., Chen, S., Scheller, H., Harholt, J., Ulvskov, P., Fox, S., Filichkin, S., Fahlgren, N., Kimbrel, J., Chang, J., Sullivan, C., Chapman, E., Carrington, J., Mockler, T., Bartley, L., Cao, P., Jung, K., Sharma, M., Vega-Sanchez, M., Ronald, P., Dardick, C., De Bodt, S., Verelst, W., Inzé, D., Heese, M., Schnittger, A., Yang, X., Kalluri, U., Tuskan, G., Hua, Z., Vierstra, R., Garvin, D., Cui, Y., Ouyang, S., Sun, Q., Liu, Z., Yilmaz, A., Grotewold, E., Sibout, R., Hematy, K., Mouille, G., Höfte, H., Michael, T., Pelloux, J., O’Connor, D., Schnable, J., Rowe, S., Harmon, F., Cass, C., Sedbrook, J., Byrne, M., Walsh, S., Higgins, J., Bevan, M., Li, P., Brutnell, T., Unver, T., Budak, H., Belcram, H., Charles, M., Chalhoub, B., & Baxter, I. (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon Nature, 463 (7282), 763-768 DOI: 10.1038/nature08747
Maize/Corn

The genome of the species known to most Americans as corn (Zea mays) and to biologists and Europeans as maize was published in the second half of 2009. Maize genetics has a history going back more than a century to the early work R. A. Emerson, widely considered the founder of modern maize genetics. Maize is an important crop species, and the most prominent crop species to engage in C4 photosynthesis (as opposed to the more standard C3 photosynthesis). The role of maize as an important model system as well as a vital crop might have placed it earlier in the order of plants to have their genomes sequenced if not for the complexity of the genome itself.
The ancestor of maize went through a Whole genome duplication between 5 and 12 million years ago. In additio, the recent history of maize has included not one but two blooms of transposon activity. The result is a genome that weighs in at ~2.5 gigabases of mostly repetitive sequence, making both sequencing and assembly major challenges.
But the maize genome sequence is now published.
The v1 sequence contains 2.3 gigabases of sequence data. Rather than shotgun sequencing of the entire genome as is now common with smaller less repetitive genomes, maize was sequenced using a BAC[5] by BAC approach. The BACs were lined up to cover the ten chromosomes of maize, and then the sequence contained in each BAC was shotgun sequenced and assembled into contigs. What this means in practice is that a given sequence in the maize genome is usually within 300 kilobases of its correct location, but within that range may be out of order or inverted. If a gene seems to be absent from its syntenic location (or only a portion of the gene is found) it is important to search up to 500 kilobases in either direction around its expected location to make sure the apparent deletion isn't the result of incorrect ordering of the contigs.
This issue was reduced in version 2 of the genome released in the spring of 2010 as over 80% of the contigs in the v2 sequence have data on their order and orientation, up from ~30% in the v1 release.
A word on gene models:
The maize genome was published with two sets of genome annotations, the working gene set and the filtered gene set. These two sets are based on different compromises between catching all the real genes in maize and excluding false genes.
- The filtered gene set (>32,000 genes (this number is from version 1, there are more in the version 2 filtered gene set released in February 2011)) are high confidence genes. If it's in the filtered gene set, it's almost certainly a gene, but there is no promise that EVERY real gene is in the filtered gene get
- The working gene set (~100,000 genes) includes all the genes in the filtered gene set, but also many other gene models that have less supporting gene evidence. Almost every real gene is likely included in the working gene set, but so are many things that aren't genes, particularly gene fragments remaining from the maize whole genome duplication, and pieces of genes captured by transposons.
The maize genome is divided among 10 chromosomes.
Maize Resources:
Maize Related CoGepedia Pages:
- Classical Maize Genes: ~460 maize genes that we have manually mapped to gene models in the published genome sequence, plus data on syntenic orthologs in rice, sorghum, and brachy, as well as the homeologous region of maize.
- MaizeGDB and CoGe: Explaining how to jump between our site and MaizeGDB
- Maize Sorghum Syntenic dotplot: How to compare the maize and sorghum genomes.
The genome paper:
Schnable, P., Ware, D., Fulton, R., Stein, J., Wei, F., Pasternak, S., Liang, C., Zhang, J., Fulton, L., Graves, T., Minx, P., Reily, A., Courtney, L., Kruchowski, S., Tomlinson, C., Strong, C., Delehaunty, K., Fronick, C., Courtney, B., Rock, S., Belter, E., Du, F., Kim, K., Abbott, R., Cotton, M., Levy, A., Marchetto, P., Ochoa, K., Jackson, S., Gillam, B., Chen, W., Yan, L., Higginbotham, J., Cardenas, M., Waligorski, J., Applebaum, E., Phelps, L., Falcone, J., Kanchi, K., Thane, T., Scimone, A., Thane, N., Henke, J., Wang, T., Ruppert, J., Shah, N., Rotter, K., Hodges, J., Ingenthron, E., Cordes, M., Kohlberg, S., Sgro, J., Delgado, B., Mead, K., Chinwalla, A., Leonard, S., Crouse, K., Collura, K., Kudrna, D., Currie, J., He, R., Angelova, A., Rajasekar, S., Mueller, T., Lomeli, R., Scara, G., Ko, A., Delaney, K., Wissotski, M., Lopez, G., Campos, D., Braidotti, M., Ashley, E., Golser, W., Kim, H., Lee, S., Lin, J., Dujmic, Z., Kim, W., Talag, J., Zuccolo, A., Fan, C., Sebastian, A., Kramer, M., Spiegel, L., Nascimento, L., Zutavern, T., Miller, B., Ambroise, C., Muller, S., Spooner, W., Narechania, A., Ren, L., Wei, S., Kumari, S., Faga, B., Levy, M., McMahan, L., Van Buren, P., Vaughn, M., Ying, K., Yeh, C., Emrich, S., Jia, Y., Kalyanaraman, A., Hsia, A., Barbazuk, W., Baucom, R., Brutnell, T., Carpita, N., Chaparro, C., Chia, J., Deragon, J., Estill, J., Fu, Y., Jeddeloh, J., Han, Y., Lee, H., Li, P., Lisch, D., Liu, S., Liu, Z., Nagel, D., McCann, M., SanMiguel, P., Myers, A., Nettleton, D., Nguyen, J., Penning, B., Ponnala, L., Schneider, K., Schwartz, D., Sharma, A., Soderlund, C., Springer, N., Sun, Q., Wang, H., Waterman, M., Westerman, R., Wolfgruber, T., Yang, L., Yu, Y., Zhang, L., Zhou, S., Zhu, Q., Bennetzen, J., Dawe, R., Jiang, J., Jiang, N., Presting, G., Wessler, S., Aluru, S., Martienssen, R., Clifton, S., McCombie, W., Wing, R., & Wilson, R. (2009). The B73 Maize Genome: Complexity, Diversity, and Dynamics Science, 326(5956), 1112-1115 DOI: 10.1126/science.1178534
Sorghum

Sorghum (Sorghum bicolor) is an important grain species. A close relative of maize, sorghum is generally considered to be an even more stress tolerant crop. Like maize it carries out C4 photosynthesis. It does not share the recent whole genome duplication seen in maize, which makes it an excellent outgroup from studies of that event in maize as the common ancestor of maize and sorghum is estimated to have lived only 12 million years ago.
The sorghum genome was published in 2009. The current version in CoGe (v1.4) contains ~700 megabases of sequence and 34,496 protein coding genes spread over ten chromosomes. The sorghum genome sequence is available from phytozome.
The genome paper: Paterson, A., Bowers, J., Bruggmann, R., Dubchak, I., Grimwood, J., Gundlach, H., Haberer, G., Hellsten, U., Mitros, T., Poliakov, A., Schmutz, J., Spannagl, M., Tang, H., Wang, X., Wicker, T., Bharti, A., Chapman, J., Feltus, F., Gowik, U., Grigoriev, I., Lyons, E., Maher, C., Martis, M., Narechania, A., Otillar, R., Penning, B., Salamov, A., Wang, Y., Zhang, L., Carpita, N., Freeling, M., Gingle, A., Hash, C., Keller, B., Klein, P., Kresovich, S., McCann, M., Ming, R., Peterson, D., Mehboob-ur-Rahman, ., Ware, D., Westhoff, P., Mayer, K., Messing, J., & Rokhsar, D. (2009). The Sorghum bicolor genome and the diversification of grasses Nature, 457 (7229), 551-556 DOI: 10.1038/nature07723
Foxtail Millet
(genome released but unpublished)
Foxtail Millet (Setaria italica) is a C4 grass. It is the first species in Paniceae, a tribe of grasses that includes switchgrass and is sister to the Andropogoneae (the tribe that maize and sorghum belong to), to have its genome sequenced. Foxtail millet was domesticated in China is much more distantly related to maize and sorghum. JGI has released an 8x assembly of the foxtail millet genome (which you can read about and download here.) The current assembly is organized in ten pseudomolecules HOWEVER this assembly was based on the sorghum genome and will not reflect inversions and translocations that occured in the Setaria genome after the ancestors of those two species diverged. A more accurate assembly based on a Setaria genetic map is in the works.
The current version of the Setaria genome includes 406 megabases of sequence and 32,095 annotated genes.
Non-angiosperms
Physcomitrella patens
Physcomitrella patens is a moss. Mosses, along with liverworts and hornworts, make up the bryophytes a group of plants that have neither flowers nor vascular tissue. We think bryophytes still look a lot like the ancestors of all land plants, but it is important to remember that bryophytes alive today, like Physcomitrella patens, have been evolving from that common ancestor for just as long as rice or arabidopsis. ~450 million years in all three cases.
The Physcomitrella genome was published in early 2008 and consists of 480 megabases of sequence and 35,938 gene models spread over 2,106 scaffolds. (Physcomitrella has 27 chromosomes.) The genome was sequenced to a depth of 8x coverage using Sanger shotgun sequencing.
Physcomitrella resources:
- The genome is available from JGI.
- More links from mossgenome.org
The genome paper:
Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, Tanahashi T, Sakakibara K, Fujita T, Oishi K, Shin-I T, Kuroki Y, Toyoda A, Suzuki Y, Hashimoto S, Yamaguchi K, Sugano S, Kohara Y, Fujiyama A, Anterola A, Aoki S, Ashton N, Barbazuk WB, Barker E, Bennetzen JL, Blankenship R, Cho SH, Dutcher SK, Estelle M, Fawcett JA, Gundlach H, Hanada K, Heyl A, Hicks KA, Hughes J, Lohr M, Mayer K, Melkozernov A, Murata T, Nelson DR, Pils B, Prigge M, Reiss B, Renner T, Rombauts S, Rushton PJ, Sanderfoot A, Schween G, Shiu SH, Stueber K, Theodoulou FL, Tu H, Van de Peer Y, Verrier PJ, Waters E, Wood A, Yang L, Cove D, Cuming AC, Hasebe M, Lucas S, Mishler BD, Reski R, Grigoriev IV, Quatrano RS, Boore JL. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319 (5859):64-9 DOI: 10.1126/science.1150646
Selaginella moellendorffii
Genome apparently unpublished. If you have information on the Selaginella moellendorffii genome, please let us know so we can add the information!
Selaginella moellendorffii is a lycophyte, an ancient branch of the plant tree of life. Like mosses, lycophytes do not have flowers, but lycophytes do have a vascular system. Lycophytes are often grouped with ferns as vascular non-seed producing plants. Selaginella has the distinction of currently being the smallest sequenced plant genome (~110 megabases, smaller than Arabidopsis!) and having a dedicated wiki.
Less genomics related, but exciting never the less is that plants that recognizably belong to the Selaginella genus can be found for the last 335-350 million years in the fossil record. That is older than the dinosaurs!
Selaginella resources:
- Selaginella wiki
- Jo Ann Banks wrote a great review on Selaginella which can be found here.
- JGI page where you can download the v1.0 assembly of the Selaginella genome.
According to the Selaginella wiki the manuscript was submitted in August of 2009, but as far as I can tell has not yet been published. (Please contact me if you know more!)
Planned, In-progress, and Private genome sequencing efforts (a partial list)
- The sunflower genome project was just announced in early 2010. While it's far too early to predict when this genome will be released, it is still worth mentioning, because species within the sunflower genus (Helianthus) have genome sizes around 3000 megabases (sometimes substantially more) making this genome an candidate to steal from maize/corn the position of largest sequenced plant genome. More information here (warning this is a pdf formatted press release)
- Bayer CropScience announced they have a complete genome sequence for canola (Bassica napus) as well as varieties of Brassica rapa and Brassica oleracea. These aren't being released publicly, but from what I've heard they are open to collaborating with individual researchers who want access to the data.
- Several different companies have announced that they have sequenced the genome of the oil palm, but to the best of our knowledge none of these sequences are publicly available. News reports of the sequencing:
- Seed plant genomes listed by JGI (the Joint Genome Institute, part of the US Department of Energy) in approximate order of progress (as best I can tell I've listed those closest to completion at the top):
- Cotton (Gossypium raimondii)
- Boechera stricta Arabidopsis relative
- Rose Gum Tree (Eucalyptus grandis)
- Thellungiella halophila Salt tolerant arabidopsis relative
- Capsella rubella Arabidopsis relative
- Seagrass (Zostera marina) a monocot.
- Brassica rapa Turnips and other vegetables
- Loblolly Pine (Pinus taeda)
- Arabidopsis halleri Arabidopsis species
- Boechera holboellii Arabidopsis relative
- Miscanthus giganteus a biofuel crop not unlike switchgrass
- Panic grass (Panicum hallii) a switchgrass relative
- Beans (Phaseolus vulgaris)
- Duck weed (Spirodela polyrhiza) a monocot
- Arabidopsis arenosa an arabidopsis species
- Boechera divericarpa an arabidopsis relative
- Switchgrass (Panicum virgatum)
- Purple willow (Salix purpure)
- Fruit tree genomes currently in the process of being sequenced/assembled/annotated by researchers including Amit Dhingra's group at Washington State:
- Pear (Pyrus communis (?) )
- Sweet Cherry (Prunus avium)
- A group of researchers are currently in the process of sequencing the genome of Common Milkweed (Asclepias syriaca) using Illumina short read sequencing.
- Other Genomes in progressCulled from abstracts from the 2011 Plant and Animal Genome Conference
- Barley
- African Cultivated Rive (Oryza glaberrima)
- Coffee (Coffea caneophora)
- Thellungiella parvula Extremophile relative of Arabidopsis thaliana
- Lettuce
- Watermelon
- Chinese Chestnut (Castanea mollissima)
- European poplar (Populus nigra)
- European aspen (Populus tremula)
- European plum (Prunus domestica)
- Red Raspberry (Rubus idaeus cv. Heritage)
- Sangiovese Grapevine (Vitis vinifera var Sangiovese)
- Peanut (Arachis hypogaea)
- Peanut (Arachis hypogaea)
Footnotes
- ↑ 1.0 1.1 Groupings of dna sequence that correspond to the individual chromosomes of an organisms
- ↑ 2.0 2.1 Need to define gene models in tomato entry
- ↑ Literally one uping the 1000 genome project that plans to sequence the genomes of 1000 people
- ↑ Estimated to be 6.7 billion people as of early 2010
- ↑ Bacterial artificial chromosome. A way of break down a genome into managable chunks of ~300 kilobases