Genomic Rearrangement Analysis
Using CoGe to help with Genomic Rearragement Analyses is quite simple:
- Pre-configured SynMap example: http://genomevolution.org/r/2nd9
- Go to SynMap
- Select two genomes
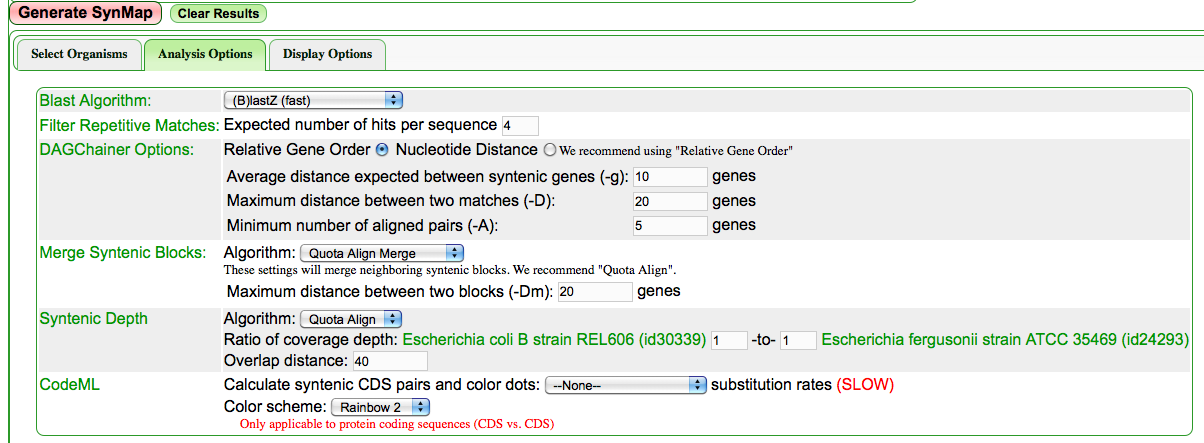
- Select the "Analysis Options" tab
- Optional: Set the "Merge Syntenic Blocks" algorithm to [[Quota align] and set some number of allowable overlapping genes (10-20 works well for microbes, select 80 for large animal and plant genomes)
- Set the "Syntenic Depth" algorithm to Quota align and have the ratio of syntenic depth set to 1:1
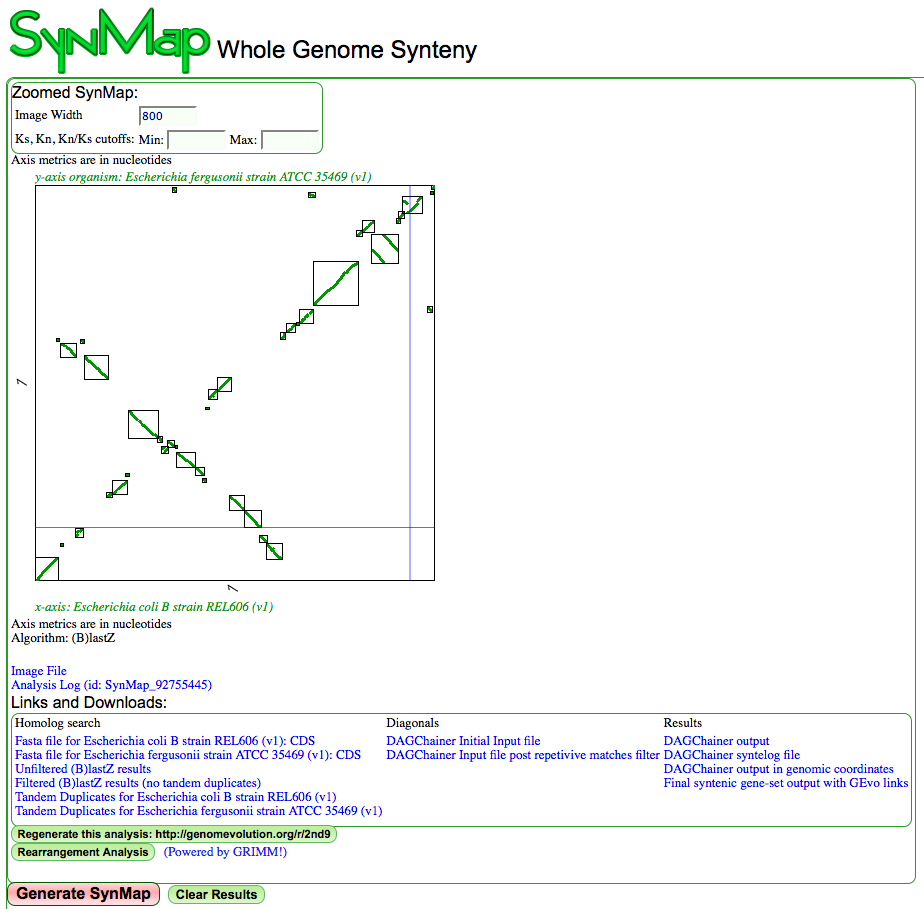
- Run SynMap
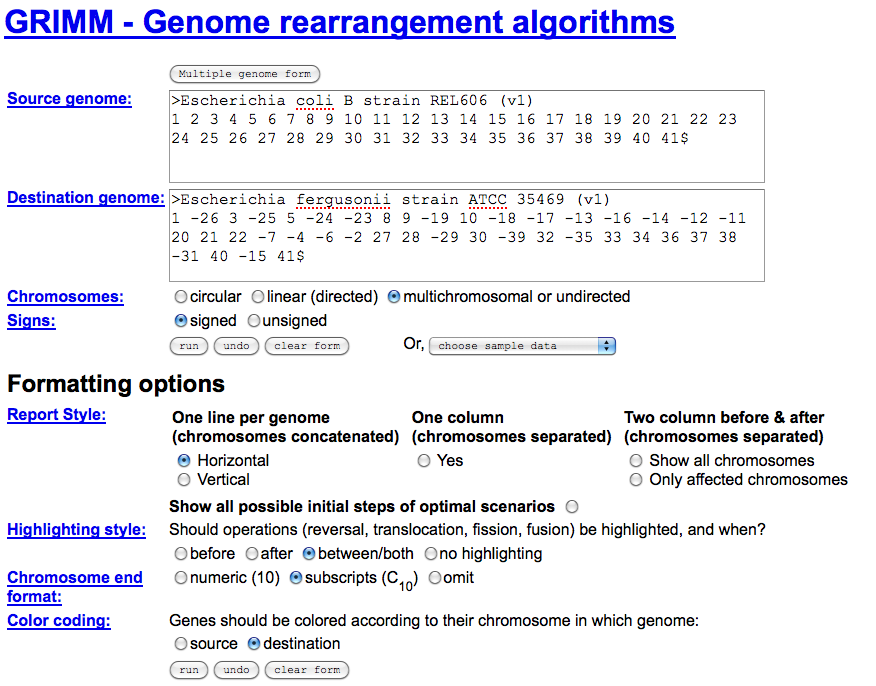
- Use the link "Rearrangement Analysis" located under the "Links and Downloads". This will take you to GRIMM for the genomic rearrangement analysis
- Run GRIMM