Pseudoassembly
Definition
Pesudoassembly is the process of creating a fake assembly of a genome by tiling contigs of a draft genome assembly against a distantly related high-quality (reference) genome using synteny. The overall structure of a pseudoassembly is based on the structure of the reference genome, and may not reflect the actual structure of the genome being pseudoassembled.
CoGe's tool SynMap has an algorithm built in to order and orient contigs of one genome against another genome. This is known as a syntenic path assembly. When this is used in SynMap, the results will contain a link to print out a pseudoassembly. Each contig is "glued" to its neighboring contig using 100 Ns (the ambiguous nucleotide).
Example
Syntenic path assembly and pseudoassembly syntenic dotplots of the Cannabis sativa (marijuana) v. Prunus persica (peach) using SynMap
-
Syntenic dotplot by [SynMap]. X-axis Cannabis sativa (marijuana); Y-axis Prunus persica (peach). Results may be regenerated: http://genomevolution.org/r/3wz3
-
Syntenic dotplot by [SynMap]. X-axis Cannabis sativa (marijuana); Y-axis Prunus persica (peach). The Cannabis genome was pseudoassembled against the peach genome using SynMap's Syntenic path assembly algorithm. The assembled genome was reloaded into CoGe and compared against the peach genome. Results may be regenerated: http://genomevolution.org/r/3x1o
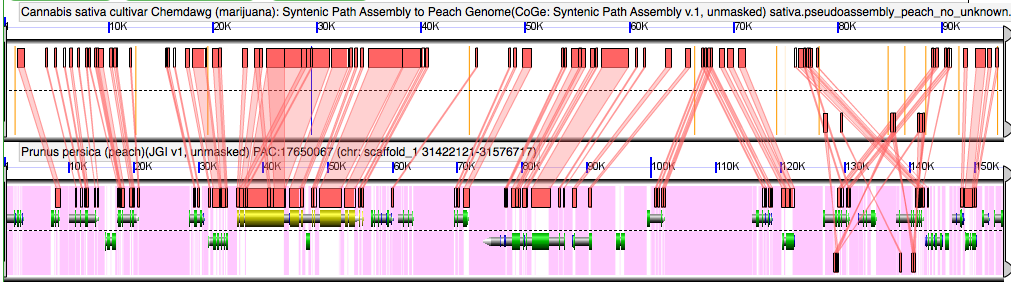
GEvo analysis of pseudoassembly of Cannabis genome to peach
Note that the pseudoassembled contigs in Cannabis are identifiable by orange bars in the background of the Cannabis panel. These orange bars are the 100 Ns used to join neighboring contigs. Results may be regenerated at http://genomevolution.org/r/3x1u .
![Syntenic dotplot by [SynMap]. X-axis Cannabis sativa (marijuana); Y-axis Prunus persica (peach). Results may be regenerated: http://genomevolution.org/r/3wz3](/wiki/images/thumb/f/f5/Master_12243_8400.genomic-CDS.lastz.dag.go_c4_D20_g10_A2.aligncoords.gcoords_ct0.w500.ass2.cs1.csoS.nsd.png/67px-Master_12243_8400.genomic-CDS.lastz.dag.go_c4_D20_g10_A2.aligncoords.gcoords_ct0.w500.ass2.cs1.csoS.nsd.png)
![Syntenic dotplot by [SynMap]. X-axis Cannabis sativa (marijuana); Y-axis Prunus persica (peach). The Cannabis genome was pseudoassembled against the peach genome using SynMap's Syntenic path assembly algorithm. The assembled genome was reloaded into CoGe and compared against the peach genome. Results may be regenerated: http://genomevolution.org/r/3x1o](/wiki/images/thumb/5/54/Master_12259_8400.genomic-CDS.lastz.dag.go_c4_D20_g10_A5.aligncoords.gcoords_ct0.w500.cs1.csoN.nsd.png/68px-Master_12259_8400.genomic-CDS.lastz.dag.go_c4_D20_g10_A5.aligncoords.gcoords_ct0.w500.cs1.csoN.nsd.png)