Qteller
qTeller is a web interface that allows researchers to extract information on genes within an interval and visualize the expression of genes as measured in multiple RNA-seq experiments. By downloading raw sequence data from multiple experiments and piping it all through the same analysis system, it is possible to generate comparable measurements of gene expression from a wide range of tissues, environmental conditions, and mutants. <-- subject always to the caveat that difference in gene expression between different studies may also be explained simply by the fact that plants were grown in different environments.
How Expression Data Is Measured
Reads are aligned to the genome using GSNAP which allows spliced alignments between multiple exons. Format conversion and alignment sorting is carred out using SAMtools and the expression of each annotated gene is quantified using Cufflinks.
Species Studied
Dedicated instances of qTeller are available for maize (25 datasets) and arabidopsis (14 datasets), however the modular nature of qTeller makes it easy to deploy new instances for any species with enough RNA-seq data to make comparative RNA-seq analysis informative.
Example Graphs
Bar Chart
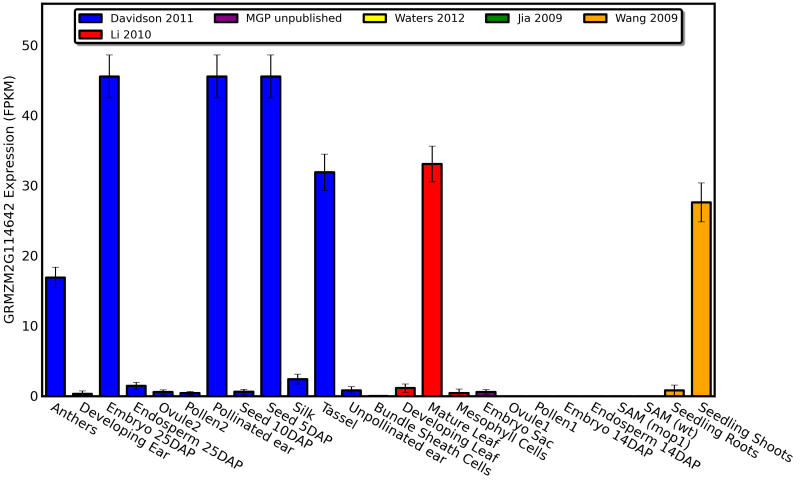
Regenerate this particular analysis by clicking this link
Scatter Plot
