Angisperm ancestral genome reconstruction
Collaboration of:
- Chunfang Zheng
- David Sankoff
- Haibao Tang
- Eric Lyons
Overview
Zheng and Sankoff have developed and continue to refine a mathematical framework for reconstructing ancestral genome states of various angiosperm lineages using syntenic blocks identified by SynMap, an interactive tool for the pairwise comparison of whole genomes development by Tang and Lyons. This problem is complicated by whole genome duplications, a common occurrence in angiosperm lineages, followed by fractionation of duplicated gene content.
Of particular interest are the ancestral genomes of:
- Eudicots which have undergone a paleohexaploidy
- Monocots
- Not possible to solve with the current taxonomic sampling of monocot genomes (currently, only grass genomes are available and of sufficient quality)
- Anticipated high-quality genomes that may help:
- Banana: http://www.musagenomics.org/
- Duckweed: http://lemna.rutgers.edu/
- Angiosperms
- Of current research interest by this team
Results
Warning: The results published here do not constitute a peer-reviewed publication. These results are a combination of work-in-progress, research notes, and various hypotheses. As such, please do not republish them, reference them, or use them as a basis for truth. However, please feel free to contact any of the researchers involved with this project if you have any questions or would like to discuss any ideas. Most of them are friendly.
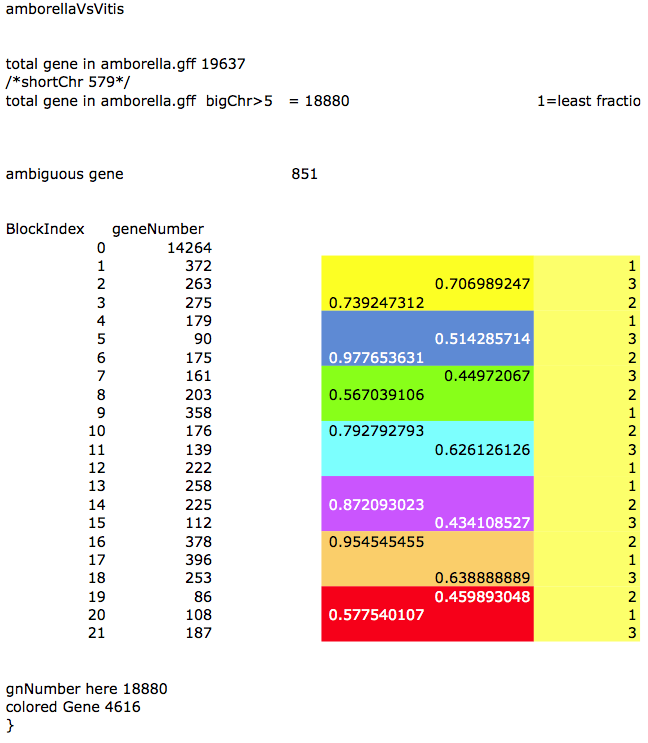
Mapping the seven hypothesized ancestral chromosomes of the pre-hexaploid onto the genome of Vitis vinifera