Duckweed Genome GC bias assessment
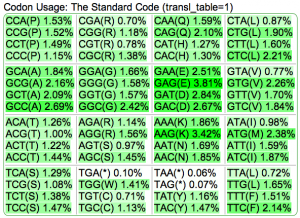
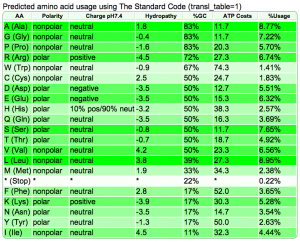
Summary: Duckweed's CDS sequences are GC biased, while the rest of the non-CDS genome is AT biased.
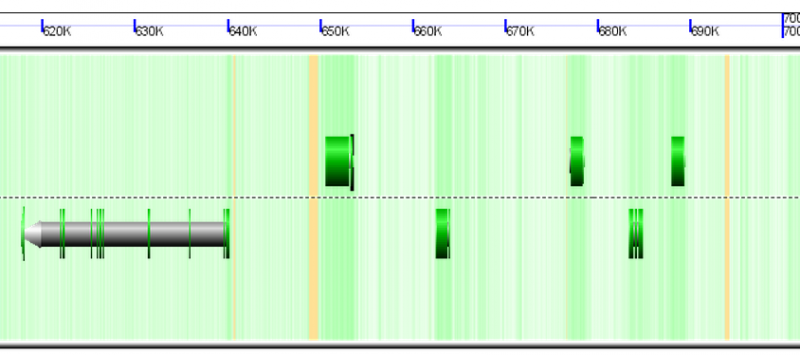
| Organism | Genome GC content | CDS GC content histogram | Codon usage | Amino acid usage |
| Duckweed | GC: 36.00% AT: 49.66% N: 14.34% X: 0.00% | 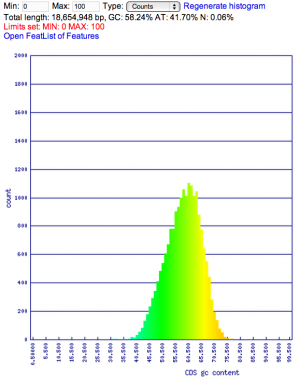 |
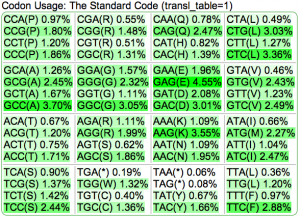 |
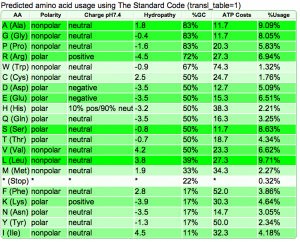 |
| Rice: http://genomevolution.org/CoGe/OrganismView.pl?dsgid=16888 | GC: 43.55% AT: 56.41% N: 0.04% X: 0.00% | 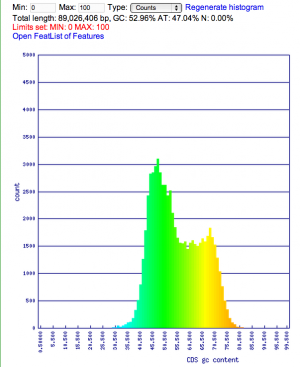 |
 |
 |