Duckweed v. Duckweed
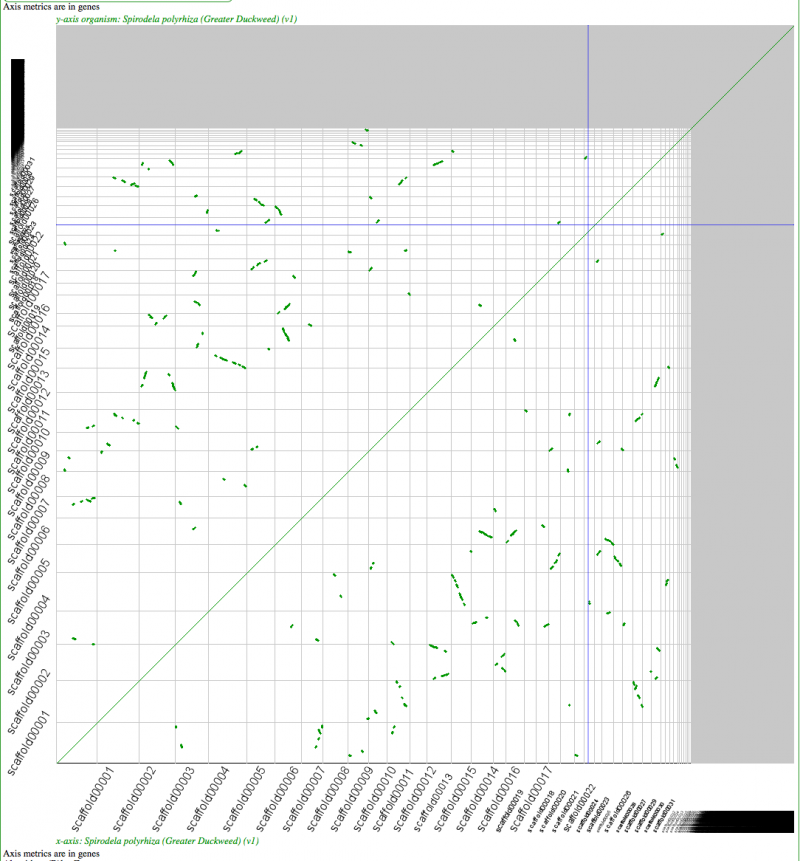
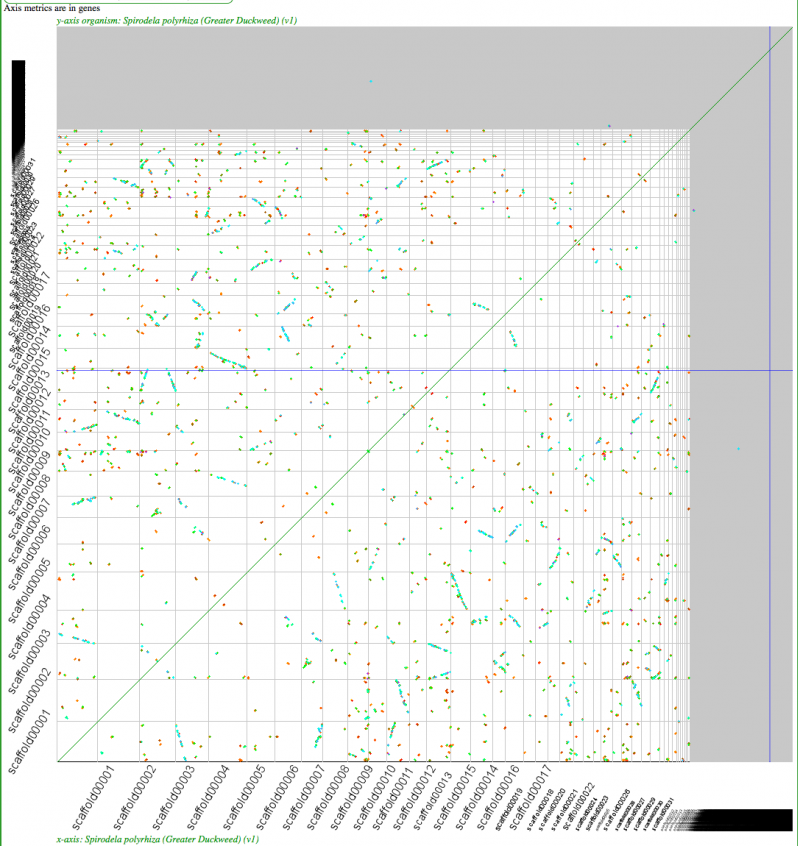
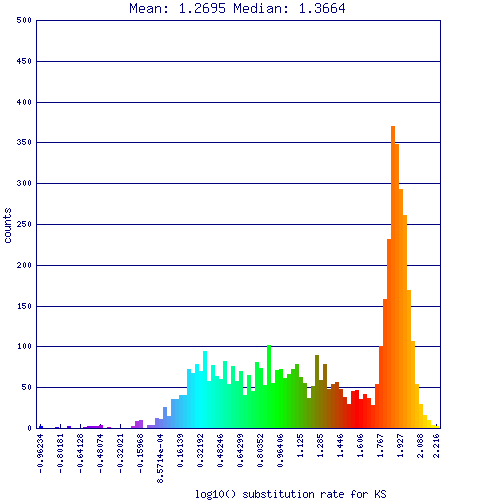
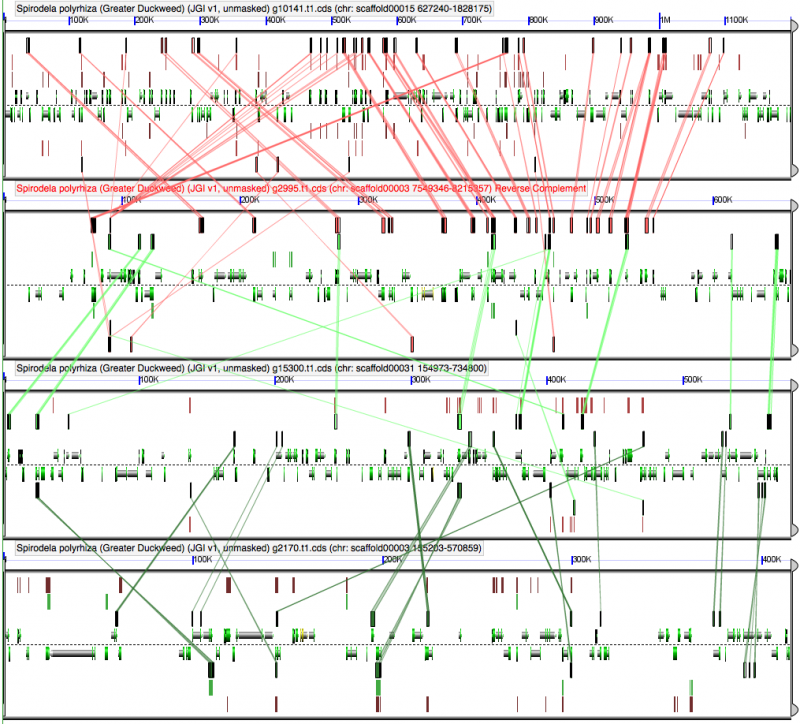
Self-self sytnenic analyses of duckweed has strong evidence for at least one whole genome duplication event. In addition, this may be a hexaploidy event or two sequential tetraploidy events (most likely two tetraploidy events). The Ks value estimates of syntenic gene pairs, which may be used to differentiate sequential whole genome duplication events (as seen in Arabidopsis), does not work with Duckweed. This is likely due to the high GC content of its coding sequence. If duckweed has undergone a recent GC content shift, then all synonymous substitution rate estimates are suspect as not being neutral. As such, Ks values does not yield a reliable metric for differentiating two sequential tetraploidy events in duckweed.