LoadExperiment
LoadExperiment enables you to load a set of experimental quantitative, polymorphism, or alignment data for a genome in CoGe. Several different file formats are supported. The data can then be viewed alongside annotation in GenomeView.
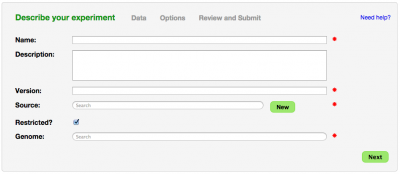
Inputs
Metadata
- Name: Name of experiment
- Description: Description of experiment
- Version: Version of experiment
- Source: Where is the data from? This could be you, your lab, your university, a sequencing center, your collaborator.
- Restricted: Is this experiment public or restricted to you and your collaborators
- Genome: Select the appropriate genome from CoGe
- Select Data File: Opens a window for specifying the input data file
- Note: Additional metadata about the experiment can be added as well.
- Example from an experiment loaded into EPIC-CoGe: http://genomevolution.org/CoGe/ExperimentView.pl?eid=193
- Information on providing a metadata file for bulk import: Experiment Metadata
Data File
You can select and retrieve data file located at:
- The iPlant Data Store
- An FTP server
- Your computer (Upload)
Data Formats
LoadExperiment supports several data file formats depending on the data type:
- Quantitative data
- Comma-separated (CSV) file format
- Tab-separated (TSV) file format
- BED file format
- Marker data
- GFF/GTF file format
- Polymorphism (SNP) data
- Variant Call Format (VCF) file format
- Alignment data
- BAM file format
Each of these file formats are described below in their own section. The file type can be auto-detected by LoadExperiment if the file name ends with the expected extension (.csv, .tsv, .bed, .vcf, .bam). Files can be compressed (.zip, .gz) and still have their type auto-detected (e.g., mydata.bed.gz). For non-standard file name extensions, you can select the file type from a list.
CSV File Format
This is a comma-delimited file that contains the following columns
- Chromosome (string)
- Start position (integer)
- Stop position (integer)
- Chromosome Strand (1 or -1)
- Measurement Value must be between [1-0] (real number; inclusive)
- Second Value (OPTIONAL): can store a second value such as an expect value (real number)
#CHR,START,STOP,STRAND,VALUE1(0-1),VALUE2(ANY-ANY) Chr1,11486,12316,1,0.181430277220112,7.3980806218146 Chr1,27309,28272,1,0.944373742485446,5.08225285439412 Chr1,32484,32978,1,0.328500324191726,1.97719838086201 Chr1,41942,42508,-1,0.825027233105203,6.56057592312617 Chr1,56394,57527,-1,0.183234367788511,0.795527328556531 Chr1,67705,68809,-1,0.956523086778851,5.20992343466606 Chr1,71144,72409,1,0.42955128220331,1.80604269639474 Chr1,81671,82833,1,0.626003507696723,2.77834108023821 Chr1,86467,87623,-1,0.0878653961575928,7.42843749315945
TSV File Format
Same as CSV format but with tab delimiters instead of commas.
BED File Format
Standard BED format as defined here: http://genome.ucsc.edu/FAQ/FAQformat.html#format1
Only the first six columns are used, with the "name" field ignored.
GFF File Format
Standard GFF3 format as defined here: http://gmod.org/wiki/GFF3
Only the seqid, start, end, score, strand, and attribute columns are used (column numbers 1, 4, 5, 6, 7, 9 respectively).
VCF File Format
Standard VCF 4.1 format as defined here: http://www.1000genomes.org/wiki/Analysis/Variant%20Call%20Format/vcf-variant-call-format-version-41
BAM File Format
Standard BAM format.
Bulk Loading
Please contact the CoGe Team if you have many experiments you wish to load. We will help you with the bulk loading.