Install coge
Installing CoGe on a blank Ubuntu machine.
Initial Dependencies
Run the following command:
sudo apt-get -y install {program}
where {program} is each of the following:
git mysql-server samtools ubuntu-dev-tools build-essential checkinstall gcc-multilib expat libexpat1-dev libgd2-xpm-dev build-essential njplot imagemagick graphviz apache2 lamp-server^ phpmyadmin swig ttf-mscorefonts-installer
Then, use:
sudo cpan App::cpanminus
sudo cpanm {module}
where {module} is each of the following:
AuthCas DBIx::Class DBIx::Class::Schema DBD::SQLite Data::UUID Data::GUID XML::SAX::Expat XML::Parser XML::Simple IPC::System::Simple CGI::Application::Dispatch CGI::Log GD::Graph::bars Moose ZMQ::LibZMQ3 File::Slurp DateTime URI::Split JSON JSON::XS
If cpanm is being troublesome, try
cpanm --reinstall
Create new mysql database
Dump CoGe Database schema
- File download: http://data.iplantcollaborative.org/quickshare/71b18508287f9fb0/cogetable.sql (AUTO_INCREMENT removed)
mysqldump -d -h localhost -u root -pXXXXXXX coge | sed 's/AUTO_INCREMENT=[0-9]*\b//' > cogetable.sql
Note: be sure to disable apparmour for MySQL.
Create new CoGe Database
create database coge
Initialize new coge database
mysql -u root -pXXXXXXXX coge < cogetable.sql
Populate a few entries in the feature_type table
- This is important because part of CoGe's code-base is keyed to feature_type_ids. This is done in order to improve performance of the system by using a feature_type_id to retrieve features of a particular type. An example is OrganismView which needs to find features of type "chromosome" in order to determine the size of a genome. The table loaded here contains 10 feature types
- Download file: http://data.iplantcollaborative.org/quickshare/ac8758f83c9b29b1/feature_type.sql
mysql -u root -pXXXXXXXXX coge < feature_type.sql
Create new user for new CoGe database
- Want a web-user with limited write privileges and a power user to load new data
create user 'coge'@'localhost' IDENTIFIED BY 'XXXXXX'; grant all privileges on coge.* to coge; create user 'coge_web'@'localhost' IDENTIFIED BY 'XXXXXX'; grant select on coge.* to coge_web; flush privileges;
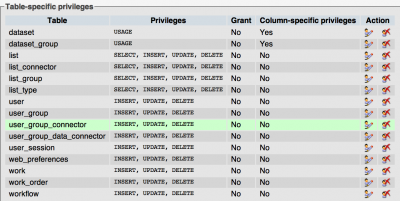
Note: The CoGe web-user needs edit/insert permission on some tables. Here is a snapshot of what these are:
Deploy new CoGe Web-site
Generate an RSS key
ssh-keygen cd .ssh
Copy the contents of the public key into Github, and then download the CoGe repository
git clone https://github.com/LyonsLab/coge.git
Make directories where files go and give write persmission
cd coge/web ./setup.sh
Configure apache
The /etc/apache2/sites-available/default.conf should look like this:
<VirtualHost *>
ServerAdmin webmaster@localhost
DocumentRoot /opt/coge/web
<Files ~ "\.(pl|cgi)$">
SetHandler perl-script
PerlResponseHandler ModPerl::Registry
Options +ExecCGI
PerlSendHeader On
</Files>
<Directory />
Options FollowSymLinks
AllowOverride None
</Directory>
<Directory /opt/coge/web>
PerlSetEnv COGE_HOME "/opt/coge/web"
Options Indexes FollowSymLinks MultiViews
AllowOverride None
Order allow,deny
allow from all
# This directive allows us to have apache2's default start page
# in /apache2-default/, but still have / go to the right place
#RedirectMatch ^/$ /apache2-default/
</Directory>
<Directory /opt/coge/web/services/>
Options +FollowSymLinks +ExecCGI
AddHandler wsgi-script .py
</Directory>
<Directory /opt/coge/web/services/JBrowse/JBrowse_TrackContent_WS/>
Options +FollowSymLinks +ExecCGI
AddHandler wsgi-script .py
</Directory>
Alias /CoGe/jex /opt/coge/web/services/jex.py
Alias /CoGe/services/JBrowse/track /opt/coge/web/services/JBrowse/J$
Alias /CoGe "/opt/coge/web"
Alias /gobe/ /opt/coge/web/gobe/
<Directory "/opt/coge/web/gobe/">
Options +FollowSymLinks +ExecCGI
AddHandler wsgi-script .py
</Directory>
ScriptALias /api/v1/jbrowse/ /opt/coge/web/services/service.pl
Alias /services/JBrowse/track /opt/coge/web/services/JBrowse/JBrowse_TrackContent_WS/source.py
ScriptAlias /api/v1/ /opt/coge/web/services/mojolicious/
ScriptAlias /cgi-bin/ /opt/coge/web/
<Directory "/opt/coge/web">
AllowOverride None
Options +ExecCGI -MultiViews +SymLinksIfOwnerMatch
AddHandler cgi-script .pl
Order allow,deny
Allow from all
</Directory>
wsgiScriptAlias /jex /opt/coge/web/services/jex.py
ErrorLog /var/log/apache2/error.log
# Possible values include: debug, info, notice, warn, error, crit,
# alert, emerg.
LogLevel warn
CustomLog /var/log/apache2/access.log combined
ServerSignature On
Alias /doc/ "/usr/share/doc/"
<Directory "/usr/share/doc/">
Options Indexes MultiViews FollowSymLinks
AllowOverride None
Order deny,allow
Deny from all
Allow from 127.0.0.0/255.0.0.0 ::1/128
</Directory>
</VirtualHost>
Configure coge.conf file
Replacing XXX's with your own information. (Change paths as necessary; this template is configured for having the Coge directory in the path: /opt/coge)
##This is a configuration file for CoGe. #database configuration DBNAME coge DBHOST localhost DBPORT 3307 DBUSER coge DBPASS XXXXXXX #CAS authentication for webservices CAS_URL https://auth.iplantcollaborative.org/cas #basic auth name and password AUTHNAME XXXXXX AUTHPASS XXXXXX #web cookie name COOKIE_NAME cogec #support email address SUPPORT_EMAIL XXXXXX #basedir for coge COGEDIR /opt/coge/web/ #bin dir for coge's programs BINDIR /opt/coge/web/bin/ #scripts dir for coge's programs SCRIPTDIR /opt/coge/scripts RESOURCESDIR /opt/coge/resources #data dir for coge's programs DATADIR /storage/coge/data/ #cache dir CACHEDIR /storage/coge/data/cache/ #dir for pair-wise whole genome comparisons (e.g. SynMap) DIAGSDIR /opt/coge/web/data/diags/ #fasta dir FASTADIR /opt/coge/web/data/fasta/ #sequence dir SEQDIR /storage/coge/data/genomic_sequence/ #SEQDIR /opt/tmp/data/ #experiment dir EXPDIR /storage/coge/data/experiments/ #TMPL dir for CoGe's web page templates TMPLDIR /opt/coge/web/tmpl/ #temp dir for coge TEMPDIR /opt/coge/web/tmp/ #secure temp dir SECTEMPDIR /storage/coge/tmp/ #IRODS dir IRODSDIR /iplant/home/<USER>/coge_data IRODSENV /opt/coge/web/irodsEnv #Base URL for web-server URL / #URL for temp directory TEMPURL /tmp/ #blast style scoring matrix dirs #BLASTMATRIX /opt/coge/web/data/blast/matrix/ BLASTMATRIX /storage/coge/data/blast/matrix/ #blastable DB #BLASTDB /opt/coge/web/data/blast/db/ BLASTDB /storage/coge/data/blast/db/ #lastable DB #LASTDB /home/franka1/coge/web/data/last/db/ LASTDB /storage/coge/data/last/db/ #directory for bed files BEDDIR /opt/coge/web/data/bed/ #servername for links SERVER http://XXXXXX/ #Job Engine Server JOBSERVER localhost #Job Engine Port JOBPORT 5151 #directory for caching genome browser images IMAGE_CACHE /opt/coge/web/data/image_cache/ #maximum number of processor to use for multi-CPU systems MAX_PROC 32 COGE_BLAST_MAX_PROC 8 #True Type Font FONT /usr/share/fonts/truetype/msttcorefonts/arial.ttf #SynMap workflow tools KSCALC /opt/coge/web/bin/SynMap/kscalc.pl GEN_FASTA /opt/coge/web/bin/SynMap/generate_fasta.pl RUN_ALIGNMENT /opt/coge/web/bin/SynMap/quota_align_merge.pl RUN_COVERAGE /opt/coge/web/bin/SynMap/quota_align_coverage.pl PROCESS_DUPS /opt/coge/web/bin/SynMap/process_dups.pl GEVO_LINKS /opt/coge/web/bin/SynMap/gevo_links.pl DOTPLOT_DOTS /opt/coge/web/bin/dotplot_dots.pl #various programs BL2SEQ /usr/local/bin/legacy_blast.pl bl2seq BLASTZ /usr/local/bin/blastz LASTZ /usr/local/bin/lastz MULTI_LASTZ /opt/coge/web/bin/blastz_wrapper/blastz.py LAST_PATH /opt/coge/web/bin/last_wrapper/ MULTI_LAST /opt/coge/web/bin/last_wrapper/last.py #BLAST 2.2.23+ BLAST /usr/local/bin/legacy_blast.pl blastall TBLASTN /usr/local/bin/tblastn BLASTN /usr/local/bin/blastn BLASTP /usr/local/bin/blastp TBLASTX /usr/local/bin/tblastx FASTBIT_LOAD /usr/local/bin/ardea FASTBIT_QUERY /usr/local/bin/ibis SAMTOOLS /usr/bin/samtools RAZIP /usr/local/bin/razip ###Formatdb needs to be updated to makeblastdb FORMATDB /usr/bin/formatdb LAGAN /opt/coge/web/bin/lagan-64bit/lagan.pl LAGANDIR /opt/coge/web/bin/lagan-64bit/ CHAOS /opt/coge/web/bin/lagan-64bit/chaos GENOMETHREADER /opt/coge/web/bin/gth DIALIGN /opt/coge/web/bin/dialign2_dir/dialign2-2_coge DIALIGN2 /opt/coge/web/bin/dialign2_dir/dialign2-2_coge DIALIGN2_DIR /opt/coge/web/bin/dialign2_dir/ HISTOGRAM /opt/coge/web/bin/histogram.pl KS_HISTOGRAM /opt/coge/web/bin/ks_histogram.pl PYTHON /usr/bin/python PYTHON26 /usr/bin/python DAG_TOOL /opt/coge/web/bin/SynMap/dag_tools.py BLAST2BED /opt/coge/web/bin/SynMap/blast2bed.pl TANDEM_FINDER /opt/coge/web/bin/dagchainer/tandems.py DAGCHAINER /opt/coge/web/bin/dagchainer_bp/dag_chainer.py EVALUE_ADJUST /opt/coge/web/bin/dagchainer_bp/dagtools/evalue_adjust.py FIND_NEARBY /opt/coge/web/bin/dagchainer_bp/dagtools/find_nearby.py QUOTA_ALIGN /opt/coge/web/bin/quota-alignment/quota_align.py CLUSTER_UTILS /opt/coge/web/bin/quota-alignment/cluster_utils.py BLAST2RAW /opt/coge/web/bin/quota-alignment/scripts/blast_to_raw.py SYNTENY_SCORE /opt/coge/web/bin/quota-alignment/scripts/synteny_score.py DOTPLOT /opt/coge/web/bin/dotplot.pl SVG_DOTPLOT /opt/coge/web/bin/SynMap/dotplot.py NWALIGN /usr/local/bin/nwalign CODEML /opt/coge/web/bin/codeml/codeml-coge CODEMLCTL /opt/coge/web/bin/codeml/codeml.ctl CONVERT_BLAST /opt/coge/web/bin/convert_long_blast_to_short_blast_names.pl DATASETGROUP2BED /opt/coge/web/bin/dataset_group_2_bed.pl ARAGORN /usr/local/bin/aragorn CLUSTALW /usr/local/bin/clustalw2 GZIP /bin/gzip GUNZIP /bin/gunzip TAR /bin/tar #MotifView MOTIF_FILE /opt/coge/web/bin/MotifView/motif_hash_dump #stuff for Mauve and whole genome alignments MAUVE /opt/coge/web/bin/GenomeAlign/progressiveMauve-muscleMatrix COGE_MAUVE /opt/coge/web/bin/GenomeAlign/mauve_alignment.pl MAUVE_MATRIX /opt/coge/web/data/blast/matrix/nt/Mauve-Matrix-GenomeAlign #newicktops is part of njplot package NEWICKTOPS /usr/bin/newicktops #convert is from ImageMagick CONVERT /usr/bin/convert #from graphviz DOT NEATO CUTADAPT /usr/local/bin/cutadapt GSNAP /usr/local/bin/gsnap CUFFLINKS /usr/local/bin/cufflinks PARSE_CUFFLINKS /opt/coge/scripts/parse_cufflinks.py GMAP_BUILD /usr/local/bin/gmap_build BOWTIE_BUILD /usr/local/bin/bowtie2-build TOPHAT /usr/local/bin/tophat #THIRD PARTY URLS GENFAMURL http://dev.gohelle.cirad.fr/genfam/?q=content/upload GRIMMURL http://grimm.ucsd.edu/cgi-bin/grimm.cgi#report QTELLER_URL http://geco.iplantc.org/qTeller
Install Perl Modules and other remaining dependencies
- Install CoGe modules /modules directory of CoGe install path
sudo perl Makefile.PL lib=/usr/local/lib/perl/5.18.2; sudo make install
- Each .pm file in /coge/web will have a list of perl modules at the top of the file. Use 'sudo cpanm' to install these.
- For each path in coge.conf that starts with /usr/local/bin, download these programs and follow the installation instructions on their respective websites.
- GMAP Gsnap, and fastbit (be sure to get version 1.3.5, rather than the most recent) require the command './configure && make && sudo make install'
- For clustalw, cufflinks, TopHat, and Bowtie (needs unzipping), copy the executable files into /usr/local/bin
- Nwalign and cutadapt need the command 'sudo python setup.py install'
After installing modules use
sudo service apache2 restart
to reset the Apache server and allow for proper testing.
Install JBrowse
Download and install the JBrowse package from http://jbrowse.org/install/
cd js unzip JBrowse-1.11.4-dev.zip mv JBrowse-1.11.4 /coge/web/js/jbrowse
Populate with test data
scripts/replicate_genome_between_coge_installations.pl -dsgid 11022 -u1 coge -p1 XXXXXXX -db1 coge -u2 oryza_coge -p2 XXXXXXX -db2 oryza_coge -sd /opt/apache/oryz_coge/data/genomic_sequence
Troubleshooting
Visualization in GEvo does not work
This relies on a system known as Gobe. Check the following things:
- Apache configuration for gobe
- Check to see if paths hard-coded into gobe/flash/service.wsgi need to be updated
- NOTE: Not sure if this is required
Working on an Atmosphere Virtual Machine
Click here for instructions on dealing with issues that occur specifically with Atmosphere Virtual machines.