Analysis of variations found in genomes of Escherichia coli strain K12 DH10B and strain B REL606 using SynMap and GEvo analysis
In this exercise you will compare the genomes of two Escherichia coli strains, K12 DH10B and B REL606, using whole genome syntenic comparison and high-resolution analyses of specific genomic regions. These analyses will use CoGe's tools SynMap and GEvo respectively, and will reveal evolutionary changes between these two genomes that happened after the divergence of their lineages. While the nucleotide sequence of these genomes is identical over large expanses of their genomes, many other types of large-scale genomic change will be discovered including phage insertions, transposon transposition, and genomic insertion, deletion, inversion, and duplication events. The computational tools used to do these analyses can be used for comparing genomes of any organisms.
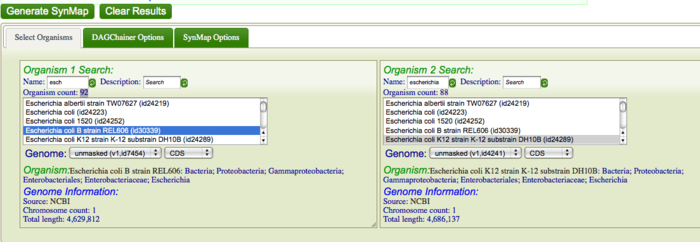
First, you are going to identify syntenic regions between these genomes. Syntenic is defined as two or more genomic regions that share a common ancestry and thus are derived from a common ancestor. To do this, you are going to construct a syntenic dotplot of K12 DH10B and B REL606 using SynMap. Go to SynMap Search for these E. coli strains in CoGe's database by typing in part of the their names in the "Name" search boxes for Organism 1 and Organism 2. For example, search for "DH10B" and "REL606" respectively, or type "escheri" in both boxes. Once CoGe has found organisms matches these names, make sure they are selected in the Organism List. While there are several parameters that can be configured when generating a syntenic dotplot using SynMap, the default settings work well for most situations, and very well for closely related organisms. Click "Generate SynMap" to start the analysis.
A lot of processing is happening behind the scenes, but the general way a syntenic dotplot is created is the genomes are compared to one another in order to find putative homologous genes between them, and then these pairs of genes are processed to find collinear series of genes in both genomes. The general principle is that the most likely and parsimonious way two genomes have a collinear series of homologous genes is those genomic regions in each organism are derived from a common ancestral genomic region (hence they are syntenic).
When finished, SynMap will display a dotplot. Each axis of the dotplot is in nucleotide units and represents one of the two genomes laid end to end. The lower-left corner represents the start of each genome (usually 'ORI' for circular bacterial genomes), and the end of each axis is the end of each chromosome. Each putative homologous gene-pair is drawn as a gray dot on the dotplot with its position corresponding to the genomic position of each gene in their respective genomes. Gene-pairs that have been identified has being syntenic are colored green. The collection of these dots appear as green line.
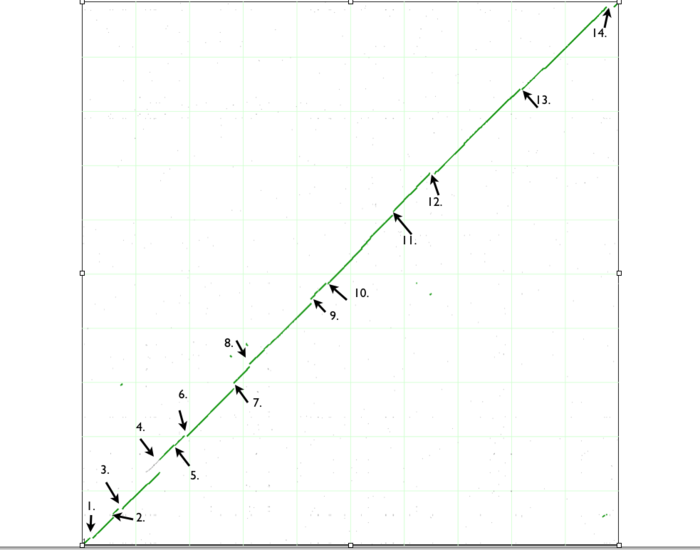
In order to accurately account for the "breaks" or discontinuities in the dotplot, we need to run GEvo analysis. GEvo uses multiple algorithms to run comparisons between the genomes. More information on GEvo software tool can be found at: GEvo. The discontinuities in our syntenic dotplot represent the sites of insertions or deletions. To analyse each of these "breaks" in dotplot, use the locator of the dotplot and place it on the green spot right before a "break". The locator will turn "red" once you have placed it on basepairs/green dots. Click after the locator turns red.
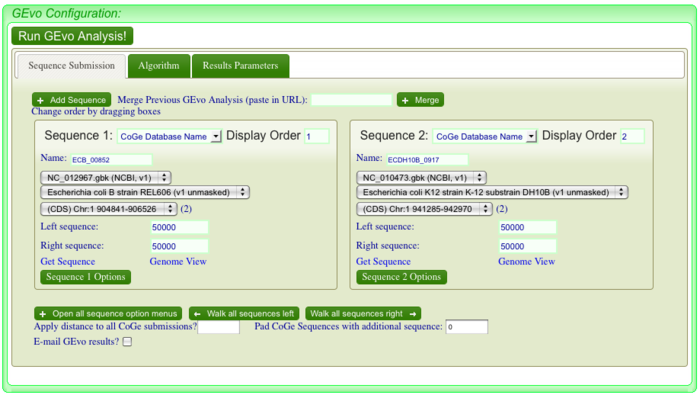
After clicking, a new page of GEvo will appear displaying the sequence information corresponding to our region of interest in the dotplot. Click "Run GEvo Analysis!". This will allow you to visualize and compare the genetic make-up of strain B REL606 and strain K DH10B at a given region. In this case, our region of interest corresponds to a discontunity in our dotplot.
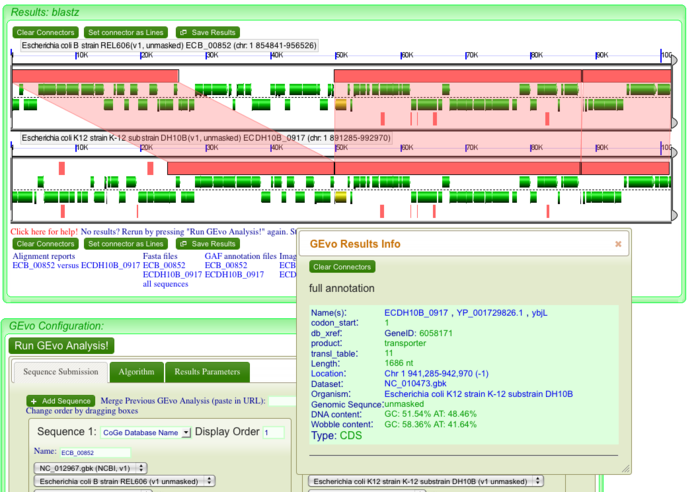
Once GEvo analysis appears, we can begin to look for differences between the two genomes. Click on individual genes/green bars and its annotation will appear in a box. Repeat the above mentioned steps for each discontinuity in dotplot for individual analysis.
. Notice the fragments of green line all over the dotplot. These represent translocation events in the E. coli strains we are examining. Place the locator on these and run GEvo to determine which genes were translocated.
Notice the pink bars over the DNA segments. Click on these and it will connect to its syntenic region. A sliding window at the sides of the diagram can be used to magnify a region and enables us to view these genes at a higher resolution. Notice the edges of pink bars connecting syntenic regions. They may run parallel or cross each other. The latter represents an inversion event.

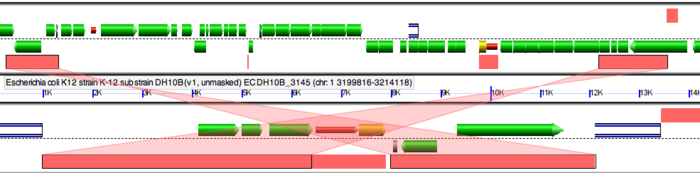
Evidence for deletion and insertions can also be found on these genomes using GEvo. At several instants, you will find that the "breaks" in our dotplot corresponds to transposition. Several deletions and insertion events could be explained by transposon activity in these genomes. The DNA segments can also be aligned against each other. This is particularly helpful when locating regions of direct repeats, inverted repeats and determining percent identities between paralogs and orthologs. To align multiple sequences simultaneously, click "+ Add Sequence", copy and paste the name/ID of the organism in the newly created box for additional sequences. Click "Run GEvo Analysis!". The resulting analysis will be color-coded distantly. Click on color-coded bars to find syntenic regions on each sequences.
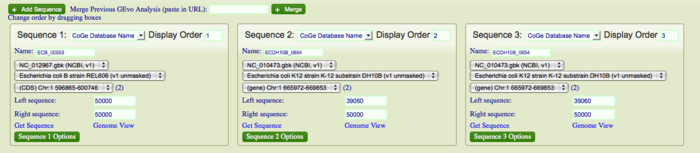
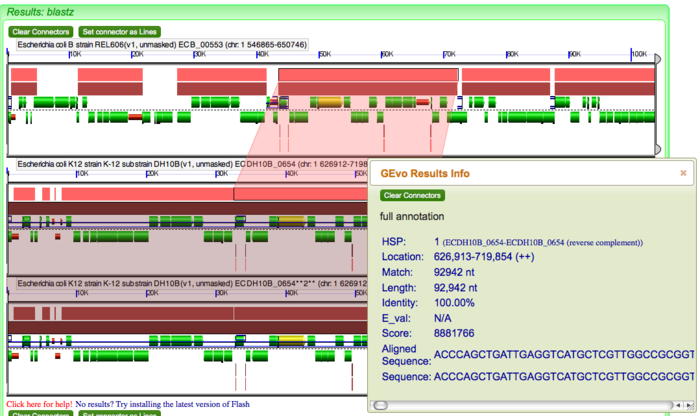
You can also distinguish the DNA segments containing different GC content relative to other parts of genomes. Under GEvo Configuration, click "Results Parameters" and select "Yes" for "Color wobble codon GC content". Click "Run GEvo Analysis!". The region containing different GC content relative to the rest of genome will be color-coded distantly.

Beware of the of the genes that may not seem syntenic (missing pink bars) at first. It is possible to assume falsely that certain genes are deleted/inserted just because not sufficient area of DNA was considered. To avoid that and locate the potential syntenic regions, change the sequence number on either genomes. Under GEvo Configuration, increase/decrease the number of sequences on left/right. Then click "Run GEvo Analysis!".
Detailed analysis of this syntenic dotplot can be found at Syntenic dotplot