Quota align: Difference between revisions
No edit summary |
|||
| (3 intermediate revisions by one other user not shown) | |||
| Line 3: | Line 3: | ||
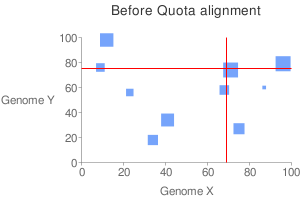
Typically in comparative genomics, we can identify anchors, chain them into syntenic blocks and interpret these blocks as derived from a common descent. However, when comparing two genomes undergone ancient genome duplications (plant genomes in particular), we have large number of blocks that are not orthologous, but are paralogous. This has forced us sometimes to use ad-hoc rules to screen these blocks. So the question is: given the expected coverage (quota) along both x- and y-axis, select a subset of the anchors with maximized total score. | Typically in comparative genomics, we can identify anchors, chain them into syntenic blocks and interpret these blocks as derived from a common descent. However, when comparing two genomes undergone ancient genome duplications (plant genomes in particular), we have large number of blocks that are not orthologous, but are paralogous. This has forced us sometimes to use ad-hoc rules to screen these blocks. So the question is: given the expected coverage (quota) along both x- and y-axis, select a subset of the anchors with maximized total score. | ||
[[Image: | [[Image:Before qa.png]][[Image:After qa.png]]<br> | ||
This program tries to screen the clusters based on the coverage constraints enforced by the user. For example, between rice-sorghum comparison, we can enforce 1:1 ratio to get all the orthologous blocks; or maybe 4:2 to grab orthologous blocks between athaliana-poplar. But the quota has to be given by the user. The program than tries to optimize the scores of these blocks globally. | This program tries to screen the clusters based on the coverage constraints enforced by the user. For example, between rice-sorghum comparison, we can enforce 1:1 ratio to get all the orthologous blocks; or maybe 4:2 to grab orthologous blocks between athaliana-poplar. But the quota has to be given by the user. The program than tries to optimize the scores of these blocks globally. | ||
For more details on the algorithm and results, please read the following article. | |||
Tang et al. (2011) ''Screening synteny blocks in pairwise genome comparisons through integer programming.'' [http://www.biomedcentral.com/1471-2105/12/102 BMC Bioinformatics] | |||
Follow this link for more information on [[syntenic depth]]. | |||
== Availability == | == Availability == | ||
The source codes are available on [http://github.com/tanghaibao/quota-alignment/ GITHUB]. Please contact haibao at berkeley dot edu for bugs/questions. | The source codes are available on [http://github.com/tanghaibao/quota-alignment/ GITHUB]. Please contact haibao at berkeley dot edu for bugs/questions. To see the algorithm in action without installation, please go to [http://genomevolution.com/CoGe/SynMap.pl CoGe SynMap tool] (also see below). Select "Analysis Options", select algorithm options for "Merge Syntenic Blocks" and/or "Syntenic Depth". | ||
== Integration with SynMap == | |||
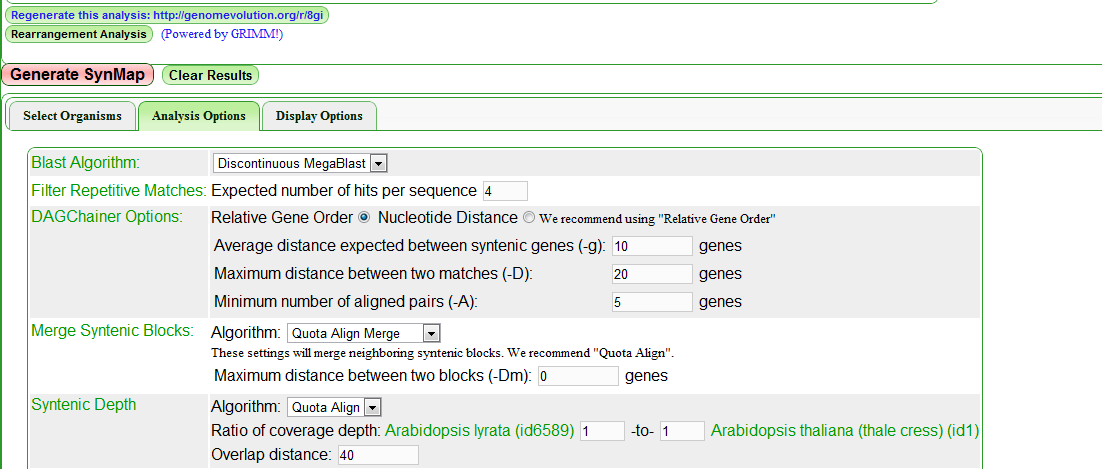
[[SynMap]] contains a set of tools within CoGe to compare genomes from many organisms and identify syntenic genomic regions. The QUOTA-ALIGN procedure can be invoked in the '''Analysis Option''' in the SynMap web interface. Users can select '''Merge syntenic blocks''' that chains syntenic blocks and '''Syntenic depth''' to specify the expected depths for screening of synteny blocks. The chaining and screening procedures are handled by QUOTA-ALIGN internally. Finally, rearrangement analysis through the [http://nbcr.sdsc.edu/GRIMM/grimm.cgi GRIMM server] is provided in the SynMap web interface.<br> | |||
[[Image:SynMap_screenshot_with_QuotaAlign.png]] | |||
The whole synteny pipeline contains a few components: 1) chaining anchors, in this case with a third-party chaining program DAGChainer; 2) 2D overlapping block merging in QUOTA-ALIGN; 3) screening of synteny blocks, in the shown example we are only interested in orthologous blocks between ''A. thaliana'' and ''A. lyrata'', and quota of 1:1 is used here for finding only the orthologous blocks; 4) encode the order for the screened blocks to [http://nbcr.sdsc.edu/GRIMM/grimm.cgi GRIMM server] for rearrangement analysis. | |||
Latest revision as of 17:22, 28 March 2012
Introduction
Typically in comparative genomics, we can identify anchors, chain them into syntenic blocks and interpret these blocks as derived from a common descent. However, when comparing two genomes undergone ancient genome duplications (plant genomes in particular), we have large number of blocks that are not orthologous, but are paralogous. This has forced us sometimes to use ad-hoc rules to screen these blocks. So the question is: given the expected coverage (quota) along both x- and y-axis, select a subset of the anchors with maximized total score.
This program tries to screen the clusters based on the coverage constraints enforced by the user. For example, between rice-sorghum comparison, we can enforce 1:1 ratio to get all the orthologous blocks; or maybe 4:2 to grab orthologous blocks between athaliana-poplar. But the quota has to be given by the user. The program than tries to optimize the scores of these blocks globally.
For more details on the algorithm and results, please read the following article.
Tang et al. (2011) Screening synteny blocks in pairwise genome comparisons through integer programming. BMC Bioinformatics
Follow this link for more information on syntenic depth.
Availability
The source codes are available on GITHUB. Please contact haibao at berkeley dot edu for bugs/questions. To see the algorithm in action without installation, please go to CoGe SynMap tool (also see below). Select "Analysis Options", select algorithm options for "Merge Syntenic Blocks" and/or "Syntenic Depth".
Integration with SynMap
SynMap contains a set of tools within CoGe to compare genomes from many organisms and identify syntenic genomic regions. The QUOTA-ALIGN procedure can be invoked in the Analysis Option in the SynMap web interface. Users can select Merge syntenic blocks that chains syntenic blocks and Syntenic depth to specify the expected depths for screening of synteny blocks. The chaining and screening procedures are handled by QUOTA-ALIGN internally. Finally, rearrangement analysis through the GRIMM server is provided in the SynMap web interface.
The whole synteny pipeline contains a few components: 1) chaining anchors, in this case with a third-party chaining program DAGChainer; 2) 2D overlapping block merging in QUOTA-ALIGN; 3) screening of synteny blocks, in the shown example we are only interested in orthologous blocks between A. thaliana and A. lyrata, and quota of 1:1 is used here for finding only the orthologous blocks; 4) encode the order for the screened blocks to GRIMM server for rearrangement analysis.