Syntenic depth
Jump to navigation
Jump to search
Syntenic depth refers to the number of times a genomic region (or genome) is syntenic to regions in another genome.
Tools in CoGe for classifying and quantitating syntenic depth:
Examples:
| Organism | Syntenic Depth |
Dotpot |
| E. coli DH10B to MG1655 |
1:1 |
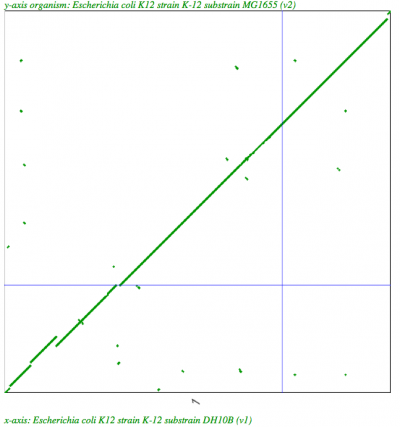 |
| Human-Chimp | 1:1 | 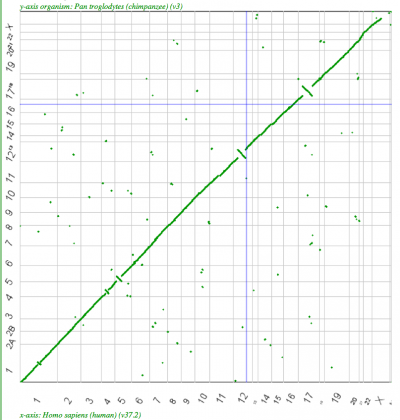
|
| Rice-Brachypodium |
2:2 |
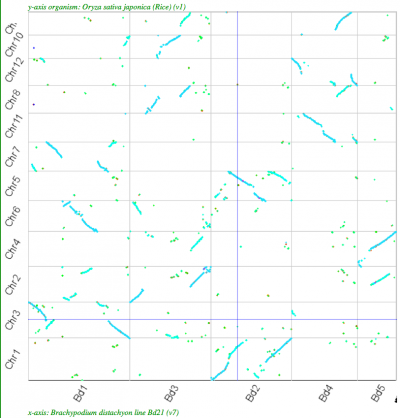 |
| Sorghum-Maize | 2:4 | 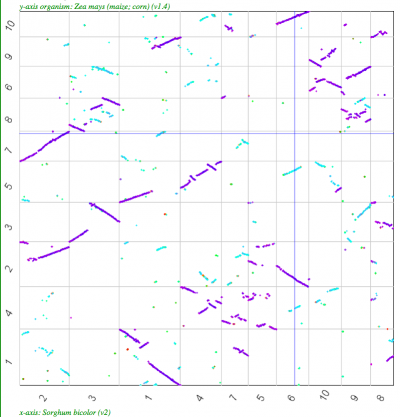
|
Syntenic Depth Tables
SynFind Syntenic Depth Examples
These tables show the number of gene participating at a particular syntenic depth and are used to infer the number of polyploidy events between two genomes.
| Syntenic Dopltot | Syntenic Depth Calculation | Known History |

|

|
Obvious 2:2 depth. Share a recent whole genome duplication. Also share older whole genome duplictions (at least one) |