Mimulus v. tomato: Difference between revisions
Jump to navigation
Jump to search
| (5 intermediate revisions by the same user not shown) | |||
| Line 6: | Line 6: | ||
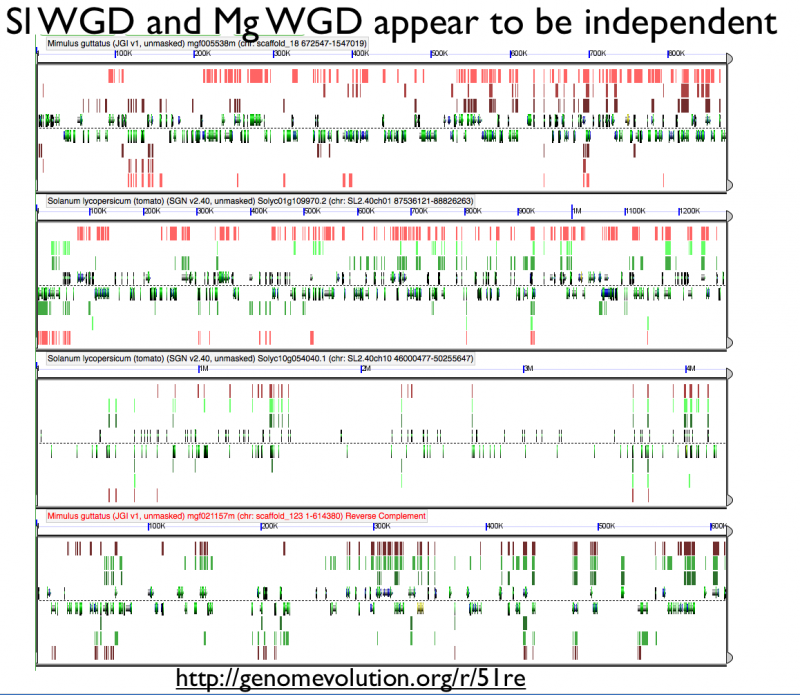
[[File:Screen shot 2012-07-06 at 3.20.46 PM.png|thumb|800px|center| http://genomevolution.org/r/51re]] | [[File:Screen shot 2012-07-06 at 3.20.46 PM.png|thumb|800px|center| http://genomevolution.org/r/51re]] | ||
[[File:Screen shot 2012-07-06 at 3.21.18 PM.png|thumb|800px|center| http://genomevolution.org/r/51mk]] | [[File:Screen shot 2012-07-06 at 3.21.18 PM.png|thumb|800px|center| http://genomevolution.org/r/51mk]] | ||
= Ks histograms of syntenic gene pairs = | |||
==Conclusion: == | |||
Mean Ks values are smaller for intragenomic comparisons than intergenomic comparisons. Evidence for independent whole genome duplications in the lineages of mimulus and solanum. | |||
===No [[syntenic depth]] screening=== | |||
{| width="800" border="1" cellpadding="1" cellspacing="1" | {| width="800" border="1" cellpadding="1" cellspacing="1" | ||
|- | |- | ||
| Line 18: | Line 21: | ||
| Mimulus guttatus | | Mimulus guttatus | ||
| Solanum lycopersicum (tomato) | | Solanum lycopersicum (tomato) | ||
| [[File:Master 8156 12289.CDS-CDS.last.dag.all.go D20 g10 A5.aligncoords.gcoords ct0.w800.ass.gene.s.ks.sr.cs1.csoS.min-0.4.log.nsd.hist.png]] http://genomevolution.org/r/51li | | [[File:Master 8156 12289.CDS-CDS.last.dag.all.go D20 g10 A5.aligncoords.gcoords ct0.w800.ass.gene.s.ks.sr.cs1.csoS.min-0.4.log.nsd.hist.png|300px]] http://genomevolution.org/r/51li | ||
| [[File:Master 8156 12289.CDS-CDS.last.dag.all.go D20 g10 A5.aligncoords.gcoords ct0.w800.ass.gene.s.ks.sr.cs1.csoS.min-0.2.max1.log.nsd.hist.png]]http://genomevolution.org/r/51up | | [[File:Master 8156 12289.CDS-CDS.last.dag.all.go D20 g10 A5.aligncoords.gcoords ct0.w800.ass.gene.s.ks.sr.cs1.csoS.min-0.2.max1.log.nsd.hist.png|300px]]]http://genomevolution.org/r/51up | ||
| | | | ||
|- | |- | ||
| Solanum lycopersicum (tomato) | | Solanum lycopersicum (tomato) | ||
| [[File:Master | | Solanum lycopersicum (tomato) | ||
| [[File:Master | | [[File:Master 12289 12289.CDS-CDS.lastz.dag.all.go D20 g10 A5.aligncoords.gcoords ct0.w1000.gene.ks.sr.cs1.csoN.log.nsd.hist.png|300px]] http://genomevolution.org/r/51uq | ||
| | | [[File:Master 12289 12289.CDS-CDS.lastz.dag.all.go D20 g10 A5.aligncoords.gcoords ct0.w1000.gene.ks.sr.cs1.csoN.min-.3.max1.log.nsd.hist.png|300px]] http://genomevolution.org/r/4tpr | ||
| | |||
|- | |- | ||
| Mimulus guttatus | |||
| Mimulus guttatus | |||
| [[File:Master 8156 8156.CDS-CDS.last.dag.all.go D20 g10 A5.aligncoords.gcoords ct0.w800.ass.gene.s.ks.sr.cs1.csoS.log.nsd.hist.png|300px]] http://genomevolution.org/r/51v0 | |||
| [[File:Master 8156 8156.CDS-CDS.last.dag.all.go D20 g10 A5.aligncoords.gcoords ct0.w800.ass.gene.s.ks.sr.cs1.csoS.min-1.max1.log.nsd.hist.png|300px]] http://genomevolution.org/r/51v1 | |||
| | |||
|} | |||
===1:1 [[syntenic depth]] screening (identified primarily orthologous syntenic gene pairs=== | |||
{| width="800" border="1" cellpadding="1" cellspacing="1" | |||
|- | |||
! scope="col" | Org 1 | |||
! scope="col" | Org 2 | |||
! scope="col" | Full Hist | |||
! scope="col" | Banded Hist | |||
! scope="col" | Notes | |||
|- | |||
| Mimulus guttatus | |||
| Solanum lycopersicum (tomato) | | Solanum lycopersicum (tomato) | ||
| [[File:Master 8156 12289.CDS-CDS.last.dag.all.go D20 g10 A5.aligncoords.qac1.1.40.gcoords ct0.w800.gene.s.ks.sr.cs1.csoS.min-.4.log.nsd.hist.png|300px]] http://genomevolution.org/r/51uv | |||
| [[File:Master | | [[File:Master 8156 12289.CDS-CDS.last.dag.all.go D20 g10 A5.aligncoords.qac1.1.40.gcoords ct0.w800.gene.s.ks.sr.cs1.csoS.min-.4.max1.log.nsd.hist.png|300px]] http://genomevolution.org/r/51uw | ||
| [[File:Master | | Ortholog screening 1:1 [[Syntenic depth]] | ||
| | |||
|- | |- | ||
| Solanum lycopersicum (tomato) | | Solanum lycopersicum (tomato) | ||
| Solanum lycopersicum (tomato) | | Solanum lycopersicum (tomato) | ||
| | | [[File:Master 12289 12289.CDS-CDS.lastz.dag.all.go D20 g10 A5.aligncoords.Dm80.ma1.qac1.1.20.gcoords ct0.w1000.gene.ks.sr.cs1.csoN.log.nsd.hist.png|300px]] http://genomevolution.org/r/51v4 | ||
| | | [[File:Master 12289 12289.CDS-CDS.lastz.dag.all.go D20 g10 A5.aligncoords.Dm80.ma1.qac1.1.20.gcoords ct0.w1000.gene.ks.sr.cs1.csoN.min-.4.max1.log.nsd.hist.png|300px]] http://genomevolution.org/r/51v6 | ||
| Ortholog screening | | Ortholog screening 1:1 [[Syntenic depth]] | ||
|- | |- | ||
| Mimulus guttatus | | Mimulus guttatus | ||
| Mimulus guttatus | | Mimulus guttatus | ||
| [[File:Master 8156 8156.CDS-CDS.last.dag.all.go D20 g10 A5.aligncoords.gcoords ct0.w800 | | [[File:Master 8156 8156.CDS-CDS.last.dag.all.go D20 g10 A5.aligncoords.qac1.1.40.gcoords ct0.w800.gene.s.ks.sr.cs1.csoS.log.nsd.hist.png|300px ]] http://genomevolution.org/r/51v5 | ||
| [[File:Master 8156 8156.CDS-CDS.last.dag.all.go D20 g10 A5.aligncoords.gcoords ct0.w800 | | [[ File:Master 8156 8156.CDS-CDS.last.dag.all.go D20 g10 A5.aligncoords.qac1.1.40.gcoords ct0.w800.gene.s.ks.sr.cs1.csoS.min-1.max1.log.nsd.hist.png|300px]] http://genomevolution.org/r/51v3 | ||
| | | Ortholog screening 1:1 [[Syntenic depth]] | ||
|} | |} | ||
<br> | <br> | ||
Latest revision as of 15:03, 7 July 2012
Note: evaluation of tomato is difficult as its polyploidy is not a string tetraploidy or hexaploidy: Tomato genome
Whole genome syntenic dotplots
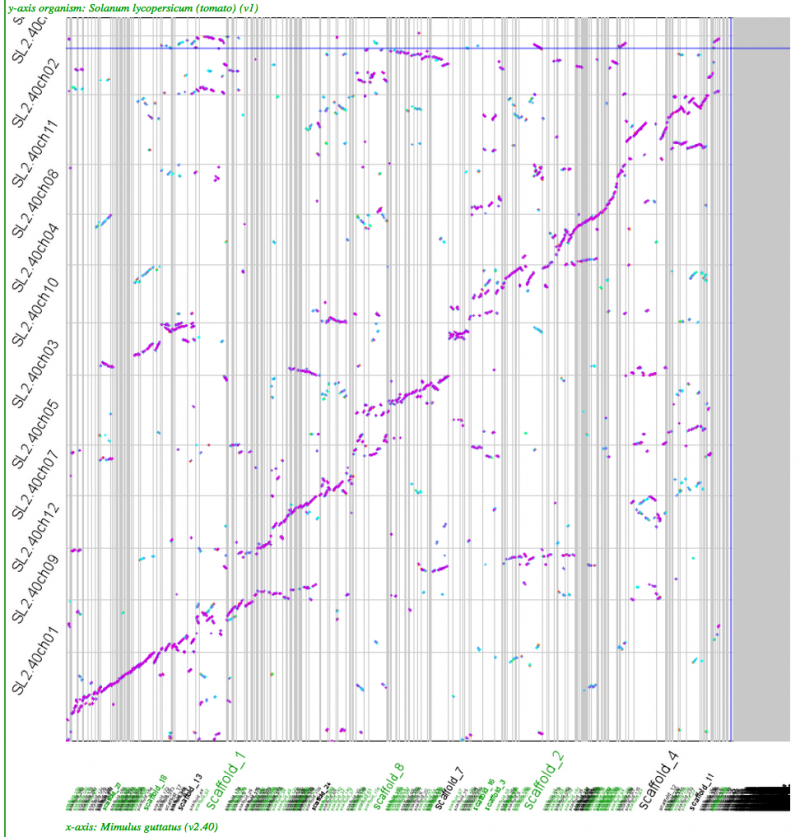
High-resolution analysis of syntenic regions with GEvo
Fractionation appears to be independent among intragenomic syntenic regions
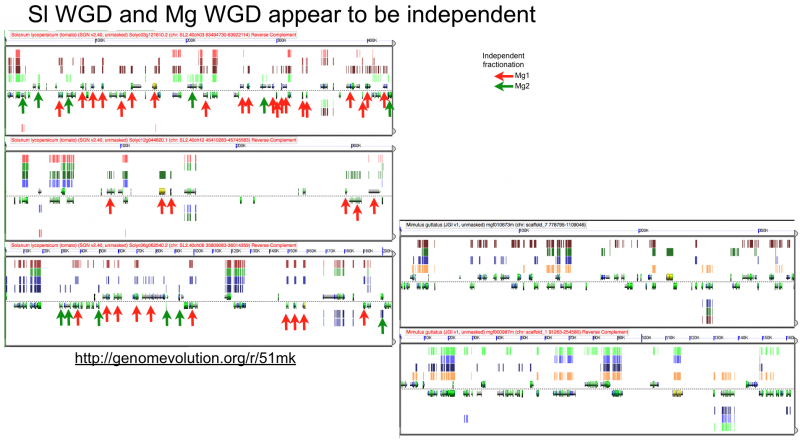
Ks histograms of syntenic gene pairs
Conclusion:
Mean Ks values are smaller for intragenomic comparisons than intergenomic comparisons. Evidence for independent whole genome duplications in the lineages of mimulus and solanum.
No syntenic depth screening
| Org 1 | Org 2 | Full Hist | Banded Hist | Notes |
|---|---|---|---|---|
| Mimulus guttatus | Solanum lycopersicum (tomato) | 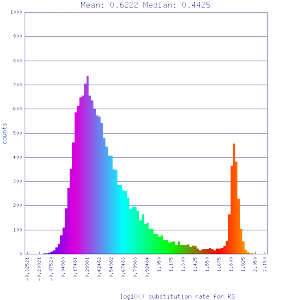 http://genomevolution.org/r/51li http://genomevolution.org/r/51li
|
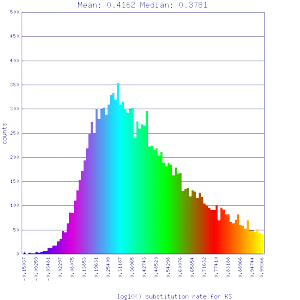 ]http://genomevolution.org/r/51up ]http://genomevolution.org/r/51up
|
|
| Solanum lycopersicum (tomato) | Solanum lycopersicum (tomato) | 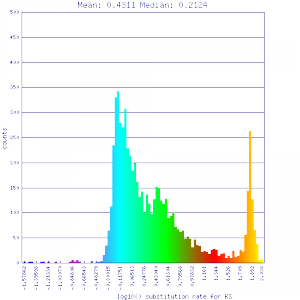 http://genomevolution.org/r/51uq http://genomevolution.org/r/51uq
|
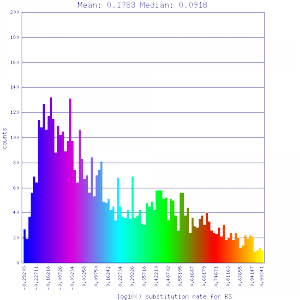 http://genomevolution.org/r/4tpr http://genomevolution.org/r/4tpr
|
|
| Mimulus guttatus | Mimulus guttatus | 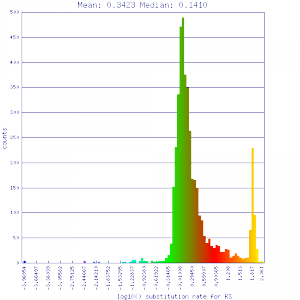 http://genomevolution.org/r/51v0 http://genomevolution.org/r/51v0
|
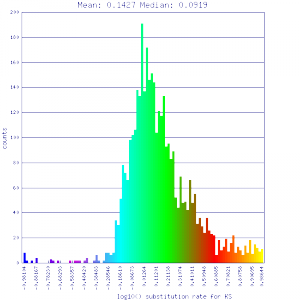 http://genomevolution.org/r/51v1 http://genomevolution.org/r/51v1
|
1:1 syntenic depth screening (identified primarily orthologous syntenic gene pairs
| Org 1 | Org 2 | Full Hist | Banded Hist | Notes |
|---|---|---|---|---|
| Mimulus guttatus | Solanum lycopersicum (tomato) | 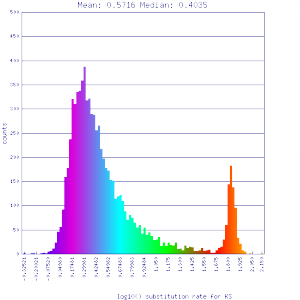 http://genomevolution.org/r/51uv http://genomevolution.org/r/51uv
|
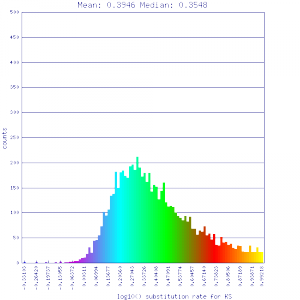 http://genomevolution.org/r/51uw http://genomevolution.org/r/51uw
|
Ortholog screening 1:1 Syntenic depth |
| Solanum lycopersicum (tomato) | Solanum lycopersicum (tomato) | 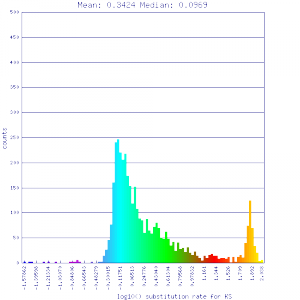 http://genomevolution.org/r/51v4 http://genomevolution.org/r/51v4
|
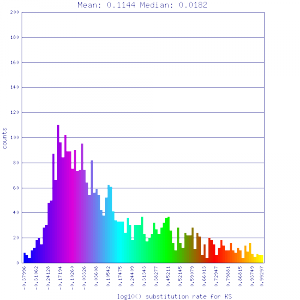 http://genomevolution.org/r/51v6 http://genomevolution.org/r/51v6
|
Ortholog screening 1:1 Syntenic depth |
| Mimulus guttatus | Mimulus guttatus | 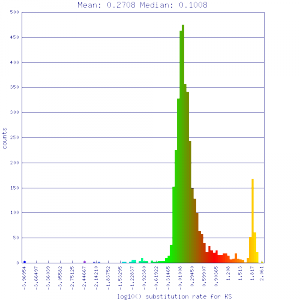 http://genomevolution.org/r/51v5 http://genomevolution.org/r/51v5
|
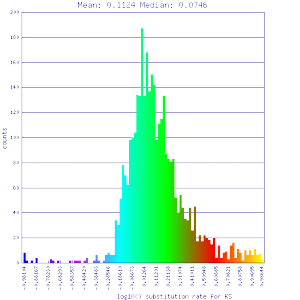 http://genomevolution.org/r/51v3 http://genomevolution.org/r/51v3
|
Ortholog screening 1:1 Syntenic depth |