Plasmodium analysis workflow 2: Tools for the syntenic analysis of whole genomes and microsyntenic regions: Difference between revisions
No edit summary |
|||
| (2 intermediate revisions by the same user not shown) | |||
| Line 10: | Line 10: | ||
===''Abstract''=== | ===''Abstract''=== | ||
Numerous genome rearrangements have taken place throughout the evolution of the genus ''Plasmodium''. These changes can be easily observed by evaluating variation on the colinearity within syntenic regions. The changes in synteny so far observed within the ''Plasmodium'' genus closely reflects species evolutionary relations. Moreover, the location of synteny breakpoints is often associated with the location of multigene families, many of which, are involved in mediating parasite-host interactions. | Numerous genome rearrangements have taken place throughout the evolution of the genus ''Plasmodium''. These changes can be easily observed by evaluating variation on the colinearity within syntenic regions. The changes in synteny so far observed within the ''Plasmodium'' genus closely reflects species evolutionary relations. Moreover, the location of synteny breakpoints is often associated with the location of multigene families, many of which, are involved in mediating parasite-host interactions. In the following sections, we explore the different CoGe tools which can be used in the study of a genome, chromosome, or species-specific rearrangement events. We assess how these events have an impact on genome architecture on both a macro and micro level. In addition, we examine genome-wide selective trends and perform preliminary genome assemblies. | ||
===''Performing synteny analyses between two genomes (SynMap)''=== | ===''Performing synteny analyses between two genomes (SynMap)''=== | ||
| Line 141: | Line 141: | ||
=== ''Using Syntenic Path Assembly (SPA) to make analysis of poor or early genome assemblies easier (SynMap - SPA tool)'' === | === ''Using Syntenic Path Assembly (SPA) to make analysis of poor or early genome assemblies easier (SynMap - SPA tool)'' === | ||
[[File:SyntenicPathAssembly.png|thumb|250px|'''Figure 8. ''P. inui'' Syntenic Path Assembly (SPA)''' using ''P. coatneyi'' as a reference genome. Black circles show putative | [[File:SyntenicPathAssembly.png|thumb|250px|'''Figure 8. ''P. inui'' Syntenic Path Assembly (SPA)''' using ''P. coatneyi'' as a reference genome. Black circles show putative inversion and duplicaion events. The analysis can be replicated following this link: https://genomevolution.org/r/ljen]] | ||
Plasmodium genomic data has markedly increased in recent years; however, there are still a large number of Plasmodium genomes that remain to be fully sequenced, assembled, and annotated. Incomplete genomic data comes from a variety of sources: | Plasmodium genomic data has markedly increased in recent years; however, there are still a large number of Plasmodium genomes that remain to be fully sequenced, assembled, and annotated. Incomplete genomic data comes from a variety of sources: | ||
| Line 150: | Line 150: | ||
Thus, tools that generate preliminary assemblies are incredibly important for ''Plasmodium'' comparative analyses. Not only can sequencing projects be streamlined by the use of a reference genome as a guide for genome assembly, but reference guided intra-specific assembly can aid in the development of species’ pan-genomes. Here, we used CoGe’s '''[[Syntenic_path_assembly]]''' ('''SPA''') tool to create a graphical display of syntenic gene pairs based on a reference genome. | Thus, tools that generate preliminary assemblies are incredibly important for ''Plasmodium'' comparative analyses. Not only can sequencing projects be streamlined by the use of a reference genome as a guide for genome assembly, but reference guided intra-specific assembly can aid in the development of species’ pan-genomes. Here, we used CoGe’s '''[[Syntenic_path_assembly]]''' ('''SPA''') tool to create a graphical display of syntenic gene pairs based on a reference genome. | ||
[[File:SPA Pvivax.png|thumb|250px|'''Figure 9.''' '''SPA''' | [[File:SPA Pvivax.png|thumb|250px|'''Figure 9.''' '''SPA''' analyses of 4 ''P. vivax'' strains. Contigs mapped to the same reference chromosome region and potential ''vir'' family members are marked with black and light red circles respectively]] | ||
[[File:SPA Pfalciparum.png|thumb|250px|'''Figure 10.''' '''SPA''' | [[File:SPA Pfalciparum.png|thumb|250px|'''Figure 10.''' '''SPA''' analyses of 4 ''P. falciparum'' strains. Contigs mapped to the same reference chromosome region are marked with purple squares]] | ||
{| class=wikitable align=center style="background: #F5FFF5;" | {| class=wikitable align=center style="background: #F5FFF5;" | ||
| Line 171: | Line 171: | ||
<span style="color:#8B008B">'''You can follow a link to an ''P. vivax'' intraspecific example analysis here:'''</span> | <span style="color:#8B008B">'''You can follow a link to an ''P. vivax'' intraspecific example analysis here:'''</span> | ||
https://genomevolution.org/r/ngvi <span style="color:#8B008B">('''Mauritania | https://genomevolution.org/r/ngvi <span style="color:#8B008B">('''Mauritania strain''')</span> | ||
https://genomevolution.org/r/ngvn <span style="color:#8B008B">('''India VII strain''')</span> | https://genomevolution.org/r/ngvn <span style="color:#8B008B">('''India VII strain''')</span> | ||
https://genomevolution.org/r/ngvf <span style="color:#8B008B">('''Brazil | https://genomevolution.org/r/ngvf <span style="color:#8B008B">('''Brazil I strain''')</span> | ||
https://genomevolution.org/r/ngwh <span style="color:#8B008B">('''North Korea strain''')</span> | https://genomevolution.org/r/ngwh <span style="color:#8B008B">('''North Korea strain''')</span> | ||
| Line 199: | Line 199: | ||
====''Genome assembly as a tool to create a P. vivax and P. falciparum pan-genome''==== | ====''Genome assembly as a tool to create a P. vivax and P. falciparum pan-genome''==== | ||
A species’ pan-genome can be used to identify common small-scale polymorphisms and larger scale indels. For instance, the ''Arabidopsis thaliana''’s pan-genome was built with specimens for a variety of geographical regions <ref>Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, Wang X, Ott F, Müller J, Alonso-Blanco C, Borgwardt K, Schmid KJ, Weigel D. 2011. Whole-genome sequencing of multiple ''Arabidopsis thaliana'' populations. Nat Genet. 43(10):956-63. https://www.ncbi.nlm.nih.gov/pubmed/21874002</ref>. As more ''Plasmodium'' genomic data become available, these intraspecific patterns of genome variability can be more clearly identified. Here, we have used '''SPA''' to identify potential intra-specific rearrangement regions and small-scale polymorphism in ''P. vivax'' ( | A species’ pan-genome can be used to identify common small-scale polymorphisms and larger scale indels. For instance, the ''Arabidopsis thaliana''’s pan-genome was built with specimens for a variety of geographical regions <ref>Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, Wang X, Ott F, Müller J, Alonso-Blanco C, Borgwardt K, Schmid KJ, Weigel D. 2011. Whole-genome sequencing of multiple ''Arabidopsis thaliana'' populations. Nat Genet. 43(10):956-63. https://www.ncbi.nlm.nih.gov/pubmed/21874002</ref>. As more ''Plasmodium'' genomic data become available, these intraspecific patterns of genome variability can be more clearly identified. Here, we have used '''SPA''' to identify potential intra-specific rearrangement regions and small-scale polymorphism in ''P. vivax'' (PO1) and ''P. falciparum'' (3D7). | ||
We analyzed 4 unassembled ''P. vivax'' strains (Brazil | We analyzed 4 unassembled ''P. vivax'' strains (Brazil I, Mauritania, North Korea, and India VII) using ''P. vivax'' (PO1) as a reference. Our results indicate that multiple contigs are syntenic to the end regions of chromosomes 1 and 6 ('''Figure 9''', black circles). In addition, patterns consistent with gene duplication are found in the central regions of chromosome 14 ('''Figure 9''', light red circles). Members of the hypervariable ''vir'' family are located at chromosome end regions, thus is possible that our findings reflect the location of strain-specific ''vir'' paralogs. | ||
Similarly, the assembly of 4 ''P. falciparum'' strains (CAMP/Malaysia, FCH/4, NF54, and Palo Alto/Uganda) shows strain-specific differences in synteny, particularly on chromosome 7 ('''Figure 10''', purple squares). Such strain-specific patterns could pinpoint the location of genomic segments of evolutionary interest. For instance, ''P. falciparum''’s AT richness and a high number of repetitive elements are factors known to negatively affect genome sequencing and assembly. Therefore, consistent assembly issues found in a specific genome region could define the location of the highly repetitive AT-rich segment, difficult to identify even by ''de novo'' assemblies. | Similarly, the assembly of 4 ''P. falciparum'' strains (CAMP/Malaysia, FCH/4, NF54, and Palo Alto/Uganda) shows strain-specific differences in synteny, particularly on chromosome 7 ('''Figure 10''', purple squares). Such strain-specific patterns could pinpoint the location of genomic segments of evolutionary interest. For instance, ''P. falciparum''’s AT richness and a high number of repetitive elements are factors known to negatively affect genome sequencing and assembly. Therefore, consistent assembly issues found in a specific genome region could define the location of the highly repetitive AT-rich segment, difficult to identify even by ''de novo'' assemblies. | ||
Latest revision as of 20:01, 15 February 2017
Outline
RETURN TO THE MAIN PAGE: Using_CoGe_for_the_analysis_of_Plasmodium_spp
RETURN TO THE PREVIOUS WORKFLOW: Plasmodium analysis workflow 1: Tools that evaluate genomic properties and amino acid usage
CONTINUE TO THE NEXT WORKFLOW: Plasmodium analysis workflow 3: Tools useful on the study of multigene families
Abstract
Numerous genome rearrangements have taken place throughout the evolution of the genus Plasmodium. These changes can be easily observed by evaluating variation on the colinearity within syntenic regions. The changes in synteny so far observed within the Plasmodium genus closely reflects species evolutionary relations. Moreover, the location of synteny breakpoints is often associated with the location of multigene families, many of which, are involved in mediating parasite-host interactions. In the following sections, we explore the different CoGe tools which can be used in the study of a genome, chromosome, or species-specific rearrangement events. We assess how these events have an impact on genome architecture on both a macro and micro level. In addition, we examine genome-wide selective trends and perform preliminary genome assemblies.
Performing synteny analyses between two genomes (SynMap)
Over evolutionary time, neighboring genes often maintain their relative position and order within a chromosomal segment. Chromosomal regions from different species that contain colinear homologs are said to be syntenic, i.e., genomic regions of shared ancestry. Changes in colinearity within syntenic regions are used to ascertain the nature, location, and extension of rearrangement events between related species. The main use of CoGE’s tool, SynMap, is to find syntenic regions where gene order is preserved. SynMap’s graphical output allows for easy and fast interpretation of these results.
| The following steps show how to analyze syntenic gene pairs with SynMap:
2. Click on Organism View or follow this link: https://genomevolution.org/coge/OrganismView.pl 3. Type a scientific name in the Search box and select the appropriate genome. Then, click on the GenomeInfo link under the Genome Information section. 4. Find the link to the SynMap tool under the Analyze section. 5. By default, SynMap will perform a self-comparison of any selected genome. This is of use when characterizing a genome or when attempting to identify the relative age of putative duplication events [1]. To analyze two different genomes, type a scientific name on the Search box of either Organism 1 or Organism 2. Once finished, click on Generate SynMap to run the analysis (Figure 1). 6. SynMap will output a graphical depiction of the syntenic regions between two genomes. There are currently two version of SynMap:
7. You can further analyze regions or genes of interest using the GEvo tool linked to SynMap. To do this, double click on a syntenic gene pair (SynMap Legacy), or select a syntenic gene pair and click on Compare in GEvo >>> (SynMap2).
https://genomevolution.org/r/lj12 (P. vivax vs. P. cynomolgi) https://genomevolution.org/r/lj1x (P. knowlesi vs. P. cynomolgi) https://genomevolution.org/r/lj1t (P. knowlesi vs. P vivax)
https://genomevolution.org/r/lq5x (P. knowlesi vs. P. malariae) https://genomevolution.org/r/lj2b (P. coatneyi vs. P. knowlesi) https://genomevolution.org/r/lq5y (P. coatneyi vs. P. malariae) https://genomevolution.org/r/lq5t (P. ovale vs. P. malariae) https://genomevolution.org/r/lq65 (P. coatneyi vs. P. ovale) https://genomevolution.org/r/lq5v (P. ovale vs. P. knowlesi) |
Identifying syntenic gene pairs
Gene position can be critical in gene expression. In many eukaryotes, expression of neighboring genes is coordinated by adjacent regulatory elements [2][3][4]. Thus, changes in gene position and order can have profound effects on gene expression. In P. falciparum, subtelomeric neighboring genes are known to form small independently expressed groups in a process thought to increase the parasite’s adaptive potential [5]. It is still unknown if these transcriptional "islands" are found outside the subtelomeric regions, or even in other Plasmodium parasites. The first step to address this issue is to use tools that allow the rapid identification of changes in gene order and position. We can use SynMap to determine gene origin, establish relative location, and identify changes in position and order. This information can later be used to establish if patterns of coordinated expression, or lack of thereof, are prevalent across the Plasmodium genus.
Identifying chromosomal inversions, fusions, fissions and other events between two genomes
Numerous genome rearrangements have taken place throughout the evolution of the genus Plasmodium. There is a strong correlation between synteny and divergence times. In other words, the more closely related two species are, the more likely synteny will be observed between their genomes [6]. We can use SynMap to identify rearrangement events and infer their putative evolutionary origin.
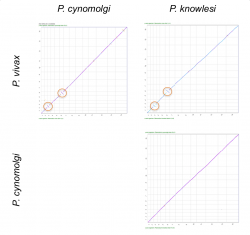
We used SynMap to confirm the location and origin of reported inversions between P. vivax, P. cynomolgi and P. knowlesi’s 3rd and 6th chromosomes. We performed pairwise comparisons to evaluate changes in genome organization among the three species (Figure 2). We only detected inversion events in pairwise comparisons with P. vivax (Figure 2, orange circles). This suggests that the inversion events reported on chromosomes 3 and 6 occurred after the split of P. cynomolgi and P. vivax (approximately 3.43-3.87 Mya) [7]. However, a detailed analysis of the breakpoint regions in P. vivax using GEvo (Figure 4) shows a genome segment of low sequence quality. Thus, it is possible that the inversion event reported on P. vivax could actually be an artifact.
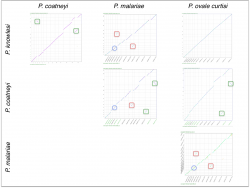
We also used SynMap to infer changes in gene order and composition among another group of closely related Plasmodium species. Pairwise comparisons were performed between four closely related Plasmodium parasites from the simian clade: P. ovale curtisi, P. malariae, P. coatneyi and P. knowlesi. We identified independent sets of chromosome fusion/fission events across these species. A set of fusions/fissions was found on P. malariae’s 5th and 9th chromosomes (Figure 3, red squares); another set of fusion/fission events was found on P. coatneyi’s 13th and 14th chromosomes (Figure 3, green squares). In addition, we found an inversion event located in the central region of P. malariae’s 4th chromosome (Figure 3, blue circle).
Measuring Kn/Ks values between genomes (SynMap - CodeML analysis tool)
Two genomes with a common ancestor will slowly accumulate nucleotide changes over time that distinguish them from one another. Nucleotide changes that result in an amino acid change are called non-synonymous and those that do not are called synonymous. Synonymous substitutions are largely neutral (have no noticeable effect) and mostly reflect background evolutionary changes. On the other hand, non-synonymous substitutions are largely affected by natural selection, as changes in a protein can give an organism a selective advantage (or be detrimental to overall fitness). Under neutrality, the rate of synonymous (Ks) and non-synonymous (Kn) substitutions will be equivalent. Deviations from this expectation indicate a significant role of natural selection. Insights into trends of natural selection are gained from evaluating the Kn/Ks ratio. We observe Kn/Ks = 1 under neutrality; we observe Kn/Ks > 1 when non-synonymous substitutions are fixed at a faster rate than synonymous ones (positive selection); and, we observe Kn/Ks < 1 when new amino acid changes are eliminated (purifying selection).
The CoGe platform is capable of calculating the Kn/Ks ratio on syntenic gene pairs across the length of a genome. CoGe’s Kn/Ks analyses can be used to:
- Identify hotspots of strong positive or purifying selection across the length of the genome.
- Establish associations between genome position (e.g. telomeres vs. centromeres) and trends of natural selection.
- Describe species- or genus-specific adaptive trends.
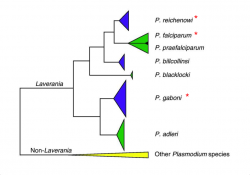
CoGe uses the CodeML analysis tool to measure the Kn/Ks ratio between two annotated genomes. The CodeML analysis tool can be accessed from SynMap. Here, we evaluated the selective trends of three closely related species from the Laveranian subgenus (Figure 18).


| The following steps show how to perform Kn/Ks analyses using SynMap’s CodeML tool:
2. Run SynMap or select a previous SynMap analysis from My Data (CoGe stores all ran analyses under a users' account). 3. Find the CodeML tool under the Analysis Options tab. Click on Calculate syntenic CDS pairs and color dots: substitution rates(s) and select Synonymous (Ks) from the dropdown menu. Repeat the analysis selecting the Non-synonymous (Kn) and (Kn/Ks) options. You can alter the display selecting a different Color Scheme, specifying Min Val. or Max Val. axis values, or changing the Log10 Transform. data option. 4. The analysis will modify the Syntenic_dotplot display to represent the distribution of the Ks, Kn or Kn/Ks values across syntenic gene pairs. A Histogram of Ks values (or Kn or Ks/Kn) will also be generated. In SynMap2, specific regions can be dynamically selected to view the Ks, Kn or Kn/Ks values.
https://genomevolution.org/r/ljhj (P. reichenowi vs. P. falciparum) https://genomevolution.org/r/ljhl (P. falciparum vs. P. gaboni) https://genomevolution.org/r/ljhq (P. reichenowi vs. P. gaboni)
https://genomevolution.org/r/lsyy (P. reichenowi vs. P. gaboni) https://genomevolution.org/r/lsz2 (P. reichenowi vs. P. falciparum) https://genomevolution.org/r/lsz5 (P. falciparum vs. P. gaboni) |
P. reichenowi and P. falciparum are thought to have diverged approximately 5.28-5.93 Mya [9]. The divergence time of either species with P. gaboni is estimated to be larger [10]. Based on these evolutionary relationships, it is expected that the number of accumulated nucleotide differences will be smaller between P. reichenowi and P. falciparum than between both species and P. gaboni.
We found smaller Ks values between P. gaboni (SY57) - P. reichenowi (CDC) than between P. gaboni (SY57) - P. falciparum (3D7) (Figure 6). Also, smaller Ks values were observed between P. reichenowi - P. falciparum than between P. falciparum - P. gaboni. The same trends were observed when a different P. reichenowi strain (SY75) was used (results can be replicated in the following links: https://genomevolution.org/r/mr5u for P. reichenowi vs. P. gaboni, and https://genomevolution.org/r/lzrr for P. reichenowi vs. P. falciparum). The differences in Ks rates suggest that a recent number of synonymous substitutions occurred on the P. reichenowi genome. Genome composition and codon usage are largely similar amongst Laveranian species (Plasmodium analysis workflow 1: Tools that evaluate genomic properties and amino acid usage). Thus, this variation could indicate an increased mutation rate in P. reichenowi, resulting in a rapidly evolving genome compared to other Laveranian. However, the reasons for this accelerated evolution remain unexplored.
Non-synonymous (Kn) substitution rates were largely similar between P. gaboni - P. falciparum and P. gaboni - P. reichenowi (Figure 7). Smaller Kn substitution values were observed between P. falciparum - P. reichenowi. Similar trends were seen when P. reichenowi (SY75) was used (results can be replicated in the following links: https://genomevolution.org/r/mr5z for P. reichenowi vs. P. gaboni, and https://genomevolution.org/r/mr5x for P. reichenowi vs. P. falciparum). These results suggest that a comparable rate of Kn changes occurred since the divergence of the P. reichenowi/P. falciparum ancestor. These changes were followed by a significant number of species-specific substitutions on both P. falciparum and P. reichenowi. Previous studies have found large Kn values in P. reichenowi - P. falciparum comparisons; particularly, in genes expressed during blood parasite's stages [11]. Thus, our results likely reflect Kn changes related to parasite-host interactions and adaptations to infection of different host types.
Using Syntenic Path Assembly (SPA) to make analysis of poor or early genome assemblies easier (SynMap - SPA tool)
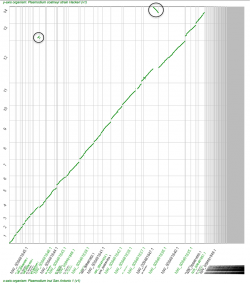
Plasmodium genomic data has markedly increased in recent years; however, there are still a large number of Plasmodium genomes that remain to be fully sequenced, assembled, and annotated. Incomplete genomic data comes from a variety of sources:
- Genomic information published on early assembly stages.
- Partially sequenced genomes.
- Low-quality genome segments.
Thus, tools that generate preliminary assemblies are incredibly important for Plasmodium comparative analyses. Not only can sequencing projects be streamlined by the use of a reference genome as a guide for genome assembly, but reference guided intra-specific assembly can aid in the development of species’ pan-genomes. Here, we used CoGe’s Syntenic_path_assembly (SPA) tool to create a graphical display of syntenic gene pairs based on a reference genome.


| The following steps show how to use SynMap - SPA tool:
2. Run SynMap between an assembled and a non-assembled genome (this might take longer than analyses using two fully assembled genomes). 3. After running SynMap click on the Display Options tab and find the SPA tool. Select the tool by clicking on the check mark next to: The Syntenic Path Assembly (SPA)?
https://genomevolution.org/r/ngvi (Mauritania strain) https://genomevolution.org/r/ngvn (India VII strain) https://genomevolution.org/r/ngvf (Brazil I strain) https://genomevolution.org/r/ngwh (North Korea strain)
https://genomevolution.org/r/ngwr (CAMP/Malaysia strain) https://genomevolution.org/r/ngxj (FCH/4 strain) https://genomevolution.org/r/ngxw (NF54 strain) https://genomevolution.org/r/ngxr (Palo Alto/Uganda strain)
|
Preliminary reference-guided assemblies
While unassembled and non-annotated genomes can be used in smaller-scale studies (e.g. orthologs can be identified with BLAST), there are limitations in their usability in large-scale comparative genomics. In order to partially circumvent this issue, we have built a preliminary assembly of the P. inui genome (on scaffold level as 2016) using the fully assembled P. coatneyi genome as a reference. Several P. inui contigs are syntenic to the same region of the reference genome (black circles, Figure 8). This result could signal a duplication event of that region on the P. inui genome or a product of the unfinished nature of the unassembled genome. However, pinpointing the event’s location using SPA could aid during de novo genome assembly. Also, contigs syntenic to a reverse DNA strand might not reflect chromosome inversions (Figure 8, black circles). Thus, while there are some limitations regarding reference-guided genome assemblies. Assembly tools such as SPA can be used to determine a preliminary location of inversions or duplications events to be later confirmed by de novo genome assemblies.
Genome assembly as a tool to create a P. vivax and P. falciparum pan-genome
A species’ pan-genome can be used to identify common small-scale polymorphisms and larger scale indels. For instance, the Arabidopsis thaliana’s pan-genome was built with specimens for a variety of geographical regions [12]. As more Plasmodium genomic data become available, these intraspecific patterns of genome variability can be more clearly identified. Here, we have used SPA to identify potential intra-specific rearrangement regions and small-scale polymorphism in P. vivax (PO1) and P. falciparum (3D7).
We analyzed 4 unassembled P. vivax strains (Brazil I, Mauritania, North Korea, and India VII) using P. vivax (PO1) as a reference. Our results indicate that multiple contigs are syntenic to the end regions of chromosomes 1 and 6 (Figure 9, black circles). In addition, patterns consistent with gene duplication are found in the central regions of chromosome 14 (Figure 9, light red circles). Members of the hypervariable vir family are located at chromosome end regions, thus is possible that our findings reflect the location of strain-specific vir paralogs.
Similarly, the assembly of 4 P. falciparum strains (CAMP/Malaysia, FCH/4, NF54, and Palo Alto/Uganda) shows strain-specific differences in synteny, particularly on chromosome 7 (Figure 10, purple squares). Such strain-specific patterns could pinpoint the location of genomic segments of evolutionary interest. For instance, P. falciparum’s AT richness and a high number of repetitive elements are factors known to negatively affect genome sequencing and assembly. Therefore, consistent assembly issues found in a specific genome region could define the location of the highly repetitive AT-rich segment, difficult to identify even by de novo assemblies.
Identifying microsyntenic regions (GEvo)

Changes in local genome organization can be used to ascertain the evolutionary history of a region (microsynteny). In Plasmodium, many genes related to parasite-host interactions are rapidly evolving and undergo frequent rearrangements, gain/loss events, and horizontal transfer. These evolutionary processes leave "genomic signals" by altering the local genome organization. Erythrocyte invasion is a multi-step process that represents one of the most crucial steps in the Plasmodium life cycle [13]. Recently, two P. falciparum genes (the reticulocyte-binding-like homologous protein 5 (Rh5) and the cysteine-rich protective antigen (CyRPA)) were shown to be the result of a horizontal gene transfer between P. faciparum and P. adleri progenitors within the . Remarkably, comparative genomics demonstrated that this horizontal gene transfer was localized to an 8kb segment on chromosome 4. The localized nature of this event, plus interspecific hybridization barriers suggest that the gene transfer occurred by the capture of a small segment of P. adleri progenitor genomic DNA by the P. falciparum progenitor within the Laveranian subgenus. As Rh5 and CyRPA are crucial for host erythrocyte invasion by P. falciparum, it has been proposed that the capture of these two genes conferred a strong fitness advantage that allowed the P. falciparum progenitor to infect humans [14]. In sum, the genomic region surrounding these two genes represents an excellent case study on how to examine microsyteny with CoGe.
Here, we will use CoGe’s tool GEvo to evaluate genomic properties within this region and assess the hypothesized horizontal transfer event.

| The following steps show how to use GEvo to analyze microsyntenic regions:
2. Click on GEvo or follow this link: https://genomevolution.org/coge/GEvo.pl 3. Specify a sequence for each box found under Sequence (you can specify a maximum of 25 sequences). Each box contains:
You can either import sequences for GEvo analysis by entering their gene IDs in the Name box, or you can select gene pairs for analysis directly from SynMap. 4. Click on Run GEvo. 5. The GEvo analysis will display the syntenic region between the compared genomes. 6. You can modify the parameters of the GEvo analysis in the Algorithm tab. Also, you can modify the information of the graphical display by altering the options on the Results Visualization Options tab.
|
We performed a microsynteny analysis of the genome region containing Rh5 and CyRPA. The analysis was conducted using the five fully sequenced Laveranian genomes currently available: P. falciparum strains 3D7 and IT, P. reichenowi strains CDC and SY57, and P. gaboni strain SY75. Our results show that microsynteny is largely maintained in the regions surrounding Rh5 and CyRPA. We modified the Results Visualization Options tab to display background and wobble GC content for genes in this region. Neither background GC content across the region, nor wobble GC content for either Rh5 or CyRPA vary significantly (Figure 11). It has been proposed that significant changes in background or wobble GC content could be used as evidence of a horizontal transfer event. However, we did not observe such a pattern in our analyses. It is possible that a horizontal transfer event between ancestral Laveranian genomes might not be detected using this method due to the similar nucleotide composition of species in the subgenus. Therefore, an additional test might be required to further support the proposed horizontal transfer event.
We also used GEvo to further analyze regions where putative inversion breakpoints are located. Comparative analyses between P. vivax (Salvador-1) and P. vivax (PO1), and between P. vivax (Salvador-1) and P. cynomolgi show two inversion events. These events are not observed in comparisons between P. cynomolgi and P. vivax (PO1). A detailed study of the inversion breakpoints using GEvo shows genome segments of low sequence quality on P. vivax (Salvador-1) (Figure 12). This suggests that the reported inversion event might be the product of a sequencing artifact instead of a real rearrangement.
Outline
RETURN TO THE MAIN PAGE: Using_CoGe_for_the_analysis_of_Plasmodium_spp
RETURN TO THE PREVIOUS WORKFLOW: Plasmodium analysis workflow 1: Tools that evaluate genomic properties and amino acid usage
CONTINUE TO THE NEXT WORKFLOW: Plasmodium analysis workflow 3: Tools useful on the study of multigene families
References
- ↑ Tang H, Lyons E. 2012. Unleashing the Genome of Brassica Rapa. Front Plant Sci. 3: 172. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3408644/
- ↑ Ghanbarian AT, Hurst LD. 2015. Neighboring Genes Show Correlated Evolution in Gene Expression. Mol Biol Evol. doi:10.1093/molbev/msv053http://mbe.oxfordjournals.org/content/early/2015/04/01/molbev.msv053.full
- ↑ De S, Teichmann SA, Babu MM. 2009. The impact of genomic neighborhood on the evolution of human and chimpanzee transcriptome. Genome Res. 19(5): 785–794. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2675967/
- ↑ Michalak P. 2008. Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics. 91:(43–248) http://www.sciencedirect.com/science/article/pii/S0888754307002807
- ↑ Rovira-Graells N, Gupta AP, Planet E, Crowley VM, Mok S, Ribas de Pouplana L, Preiser PR, Bozdech Z, Cortés A. 2012. Transcriptional variation in the malaria parasite Plasmodium falciparum. Genome Res. 5:925-38. https://www.ncbi.nlm.nih.gov/pubmed/22415456
- ↑ Tachibana SI, Sullivan SA, Kawai S, Nakamura S, Kim HR, Goto N, Arisue N, Palacpac NM, Honma H, Yagi M, Tougan T, Katakai Y, Kaneko O, Mita T, Kita K, Yasutomi Y, Sutton PL, Shakhbatyan R, Horii T, Yasunaga T, Barnwell JB, Escalante AA, Carlton JM, Tanabe K. 2012. Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nat Genet. 44: 1051–1055. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3759362/
- ↑ Pacheco MA, Reid MJ, Schillaci MA, Lowenberger CA, Galdikas BM, Jones-Engel L, Escalante AA. 2012. The origin of malarial parasites in orangutans. PLoS One. 7:e34990. https://www.ncbi.nlm.nih.gov/pubmed/22536346
- ↑ Rayner JC, Liu W, Peeters M, Sharp PM, Hahn BH. 2011. A plethora of Plasmodium species in wild apes: a source of human infection? Trends Parasitol. 27:222-9. https://www.ncbi.nlm.nih.gov/pubmed/21354860?dopt=Abstract&holding=npg
- ↑ Pacheco MA, Reid MJ, Schillaci MA, Lowenberger CA, Galdikas BM, Jones-Engel L, Escalante AA. 2012. The origin of malarial parasites in orangutans. PLoS One. 7:e34990. https://www.ncbi.nlm.nih.gov/pubmed/22536346
- ↑ Sundararaman SA, Plenderleith LJ, Liu W, Loy DE, Learn GH, Li Y, Shaw KS, Ayouba A, Peeters M, Speede S5, Shaw GM, Bushman FD, Brisson D, Rayner JC, Sharp PM, Hahn BH. 2016. Genomes of cryptic chimpanzee Plasmodium species reveal key evolutionary events leading to human malaria. Nat Commun. 7:11078. https://www.ncbi.nlm.nih.gov/pubmed/27002652
- ↑ Otto TD, Rayner JC, Böhme U, Pain A, Spottiswoode N, Sanders M, Quail M, Ollomo B, Renaud F, Thomas AW, Prugnolle F, Conway DJ, Newbold C, Berriman M. 2014. Genome sequencing of chimpanzee malaria parasites reveals possible pathways of adaptation to human hosts. Nat Commun. 5:4754. https://www.ncbi.nlm.nih.gov/pubmed/25203297
- ↑ Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, Wang X, Ott F, Müller J, Alonso-Blanco C, Borgwardt K, Schmid KJ, Weigel D. 2011. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 43(10):956-63. https://www.ncbi.nlm.nih.gov/pubmed/21874002
- ↑ Cowman AF, Crabb BS. 2006. Invasion of red blood cells by malaria parasites. Cell. 124:755-66. https://www.ncbi.nlm.nih.gov/pubmed/16497586
- ↑ Sundararaman SA, Plenderleith LJ, Liu W, Loy DE, Learn GH, Li Y, Shaw KS, Ayouba A, Peeters M, Speede S, Shaw GM, Bushman FD, Brisson D, Rayner JC, Sharp PM, Hahn BH. 2016. Genomes of cryptic chimpanzee Plasmodium species reveal key evolutionary events leading to human malaria. Nat Commun. 7:11078. https://www.ncbi.nlm.nih.gov/pubmed/27002652