Install coge: Difference between revisions
| Line 410: | Line 410: | ||
# BWAmeth: | # BWAmeth: | ||
# Homer: http://homer.salk.edu/homer/ ... sudo perl ./configureHomer.pl -install homer | # Homer: http://homer.salk.edu/homer/ ... sudo perl ./configureHomer.pl -install homer | ||
# Blat (required by Homer): | # Blat (required by Homer): https://genome.ucsc.edu/FAQ/FAQblat.html | ||
=== Install Third-Party Fonts === | === Install Third-Party Fonts === | ||
Revision as of 22:52, 7 June 2016
Installing CoGe on Ubuntu
Note: these instructions were last updated and verified on June 3rd, 2016.
Initial Dependencies
Run the following command:
sudo apt-get -y install {package}
where {package} is each of the following:
apache2 aragorn blast2 build-essential checkinstall expat gcc-multilib git graphviz imagemagick libdb-dev libgd2-xpm-dev libperl-dev libgd-gd2-perl libconfig-yaml-perl libssl-dev libzmq3-dev mysql-server ncbi-blast+ ncbi-blast+-legacy njplot phpmyadmin python-dev python-numpy python-software-properties samtools swig sqlite3 ttf-mscorefonts-installer ubuntu-dev-tools libapache-asp-perl libapache2-mod-perl2 libapache2-mod-wsgi python-pip nodejs npm libboost-all-dev (for TopHat)
Create MySQL database
Dump CoGe database schema (if using existing CoGe installation, otherwise see schema file below).
mysqldump -d -h localhost -u root -pXXXXXXX coge | sed 's/AUTO_INCREMENT=[0-9]*\b//' > coge_mysql_schema.sql
- CoGe MySQL database schema file (updated June 3rd, 2016): http://genomevolution.org/coge/coge_mysql_schema.sql
- Note: be sure to disable AppArmor for MySQL.
Create new CoGe Database
create database coge
Initialize new coge database
mysql -u root -pXXXXXXXX coge < coge_mysql_schema.sql
Populate a few entries in the feature_type table
- Use the table here which contains 10 feature types: http://genomevolution.org/coge/coge_feature_types.sql
mysql -u root -pXXXXXXXXX coge < coge_feature_types.sql
Create new MySQL user for the CoGe database
use mysql; create user 'coge'@'localhost' IDENTIFIED BY 'XXXXXX'; grant all privileges on coge.* to coge; flush privileges;
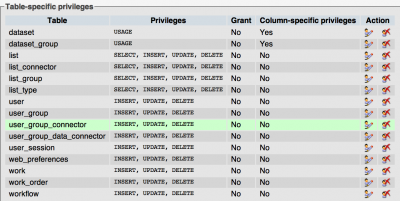
Note: The CoGe web-user needs edit/insert permission on some tables. Here is a snapshot of what these are:
Deploy the Web Site
Generate a public key and add to your GitHub account
See https://help.github.com/articles/generating-an-ssh-key/
Download the CoGe repository
git clone https://github.com/LyonsLab/coge.git
Run setup script to make required subdirectories
cd coge/web ./setup.sh
Configure apache
The /etc/apache2/sites-available/default.conf should look like this:
<VirtualHost *>
ServerAdmin webmasterl@localhost
DocumentRoot /opt/coge/web
<Files *.pl>
SetHandler perl-script
PerlResponseHandler ModPerl::Registry
Options +ExecCGI
PerlSendHeader On
</Files>
<Directory />
Options FollowSymLinks
AllowOverride None
</Directory>
Alias /gobe/ /opt/coge/web/gobe/
<Directory /opt/coge/web/gobe/>
Options +FollowSymLinks +ExecCGI
AddHandler wsgi-script .py
</Directory>
<Directory /opt/coge>
Options Includes ExecCGI FollowSymLinks
AllowOverride All
SetEnv COGE_HOME "/opt/coge/"
Order allow,deny
Allow from all
</Directory>
<Directory /opt/coge/web/services/>
Options +FollowSymLinks +ExecCGI
AddHandler wsgi-script .py
</Directory>
<Directory /opt/coge/web/services/JBrowse/JBrowse_TrackContent_WS/>
Options +FollowSymLinks +ExecCGI
AddHandler wsgi-script .py
</Directory>
ScriptAliasMatch (?i)^/coge/jex(.*) /opt/coge/web/services/jex.py/$1
AliasMatch (?i)^/coge(.*) /opt/coge/web/$1
ProxyPass /coge/api/v1/ http://localhost:3303/
ProxyPassReverse /coge/api/v1/ http://localhost:3303/
ErrorLog /var/log/apache2/error.log
# Possible values include: debug, info, notice, warn, error, crit, alert, emerg.
LogLevel warn
CustomLog /var/log/apache2/access.log combined
ServerSignature On
</VirtualHost>
Enable Required Apache Modules
sudo a2enmod rewrite headers proxy proxy_http expires perl ssl
and reset Apache
Configure coge.conf file
Replacing XXX's with your own information. (Change paths as necessary; this template is configured for having the Coge directory in the path: /opt/coge)
##This is a configuration file for CoGe. #database configuration DB mysql DBNAME coge DBHOST localhost DBPORT 3307 DBUSER coge DBPASS XXXXXXX #CAS authentication for webservices CAS_URL https://auth.iplantcollaborative.org/cas USER_API_URL https://agave.iplantc.org:443/profiles/v2 #basic auth name and password AUTHNAME XXXXXX AUTHPASS XXXXXX #web cookie name COOKIE_NAME cogec #support email address SUPPORT_EMAIL XXXXXX #basedir for coge COGEDIR /opt/coge/web/ #bin dir for coge's programs BINDIR /opt/coge/web/bin/ #scripts dir for coge's programs SCRIPTDIR /opt/coge/scripts #resources dir for static files RESOURCESDIR /opt/coge/resources #data dir for coge's programs DATADIR /storage/coge/data/ #cache dir CACHEDIR /storage/coge/data/cache/ #dir for pair-wise whole genome comparisons (e.g. SynMap) DIAGSDIR /opt/coge/web/data/diags/ #fasta dir FASTADIR /opt/coge/web/data/fasta/ #sequence dir SEQDIR /storage/coge/data/genomic_sequence/ #experiment dir EXPDIR /storage/coge/data/experiments/ #TMPL dir for CoGe's web page templates TMPLDIR /opt/coge/web/tmpl/ #temp dir for coge TEMPDIR /opt/coge/web/tmp/ #secure temp dir SECTEMPDIR /storage/coge/tmp/ #IRODS dir IRODSDIR /iplant/home/<USER>/coge_data IRODSSHARED /iplant/home/shared IRODSENV /opt/coge/web/irodsEnv #Base URL for web-server URL /coge/ #URL for temp directory TEMPURL /coge/tmp/ #blast style scoring matrix dirs BLASTMATRIX /storage/coge/data/blast/matrix/ #blastable DB BLASTDB /storage/coge/data/blast/db/ #lastable DB LASTDB /storage/coge/data/last/db/ #directory for bed files BEDDIR /opt/coge/web/data/bed/ #servername for links SERVER http://XXXXXX/ #Job Engine Server JOBSERVER localhost #Job Engine Port JOBPORT 5151 #directory for caching genome browser images IMAGE_CACHE /opt/coge/web/data/image_cache/ #maximum number of processor to use for multi-CPU systems MAX_PROC 32 COGE_BLAST_MAX_PROC 8 #True Type Font FONT /usr/share/fonts/truetype/msttcorefonts/arial.ttf #SynMap workflow tools KSCALC /opt/coge/web/bin/SynMap/kscalc.pl GEN_FASTA /opt/coge/web/bin/SynMap/generate_fasta.pl RUN_ALIGNMENT /opt/coge/web/bin/SynMap/quota_align_merge.pl RUN_COVERAGE /opt/coge/web/bin/SynMap/quota_align_coverage.pl PROCESS_DUPS /opt/coge/web/bin/SynMap/process_dups.pl GEVO_LINKS /opt/coge/web/bin/SynMap/gevo_links.pl DOTPLOT_DOTS /opt/coge/web/bin/dotplot_dots.pl #various programs BL2SEQ /usr/local/bin/legacy_blast.pl bl2seq BLASTZ /usr/local/bin/blastz LASTZ /usr/local/bin/lastz MULTI_LASTZ /opt/coge/web/bin/blastz_wrapper/blastz.py LAST_PATH /opt/coge/web/bin/last_wrapper/ MULTI_LAST /opt/coge/web/bin/last_wrapper/last.py #BLAST 2.2.23+ BLAST /usr/local/bin/legacy_blast.pl blastall TBLASTN /usr/local/bin/tblastn BLASTN /usr/local/bin/blastn BLASTP /usr/local/bin/blastp TBLASTX /usr/local/bin/tblastx FASTBIT_LOAD /usr/local/bin/ardea FASTBIT_QUERY /usr/local/bin/ibis SAMTOOLS /usr/bin/samtools RAZIP /usr/local/bin/razip ###Formatdb needs to be updated to makeblastdb FORMATDB /usr/bin/formatdb LAGAN /opt/coge/web/bin/lagan-64bit/lagan.pl LAGANDIR /opt/coge/web/bin/lagan-64bit/ CHAOS /opt/coge/web/bin/lagan-64bit/chaos GENOMETHREADER /opt/coge/web/bin/gth DIALIGN /opt/coge/web/bin/dialign2_dir/dialign2-2_coge DIALIGN2 /opt/coge/web/bin/dialign2_dir/dialign2-2_coge DIALIGN2_DIR /opt/coge/web/bin/dialign2_dir/ HISTOGRAM /opt/coge/web/bin/histogram.pl KS_HISTOGRAM /opt/coge/web/bin/ks_histogram.pl PYTHON /usr/bin/python PYTHON26 /usr/bin/python DAG_TOOL /opt/coge/web/bin/SynMap/dag_tools.py BLAST2BED /opt/coge/web/bin/SynMap/blast2bed.pl TANDEM_FINDER /opt/coge/web/bin/dagchainer/tandems.py DAGCHAINER /opt/coge/web/bin/dagchainer_bp/dag_chainer.py EVALUE_ADJUST /opt/coge/web/bin/dagchainer_bp/dagtools/evalue_adjust.py FIND_NEARBY /opt/coge/web/bin/dagchainer_bp/dagtools/find_nearby.py QUOTA_ALIGN /opt/coge/web/bin/quota-alignment/quota_align.py CLUSTER_UTILS /opt/coge/web/bin/quota-alignment/cluster_utils.py BLAST2RAW /opt/coge/web/bin/quota-alignment/scripts/blast_to_raw.py SYNTENY_SCORE /opt/coge/web/bin/quota-alignment/scripts/synteny_score.py DOTPLOT /opt/coge/web/bin/dotplot.pl SVG_DOTPLOT /opt/coge/web/bin/SynMap/dotplot.py NWALIGN /usr/bin/nwalign CODEML /opt/coge/web/bin/codeml/codeml-coge CODEMLCTL /opt/coge/web/bin/codeml/codeml.ctl CONVERT_BLAST /opt/coge/web/bin/convert_long_blast_to_short_blast_names.pl DATASETGROUP2BED /opt/coge/web/bin/dataset_group_2_bed.pl ARAGORN /usr/local/bin/aragorn CLUSTALW /usr/local/bin/clustalw2 GZIP /bin/gzip GUNZIP /bin/gunzip TAR /bin/tar #MotifView MOTIF_FILE /opt/coge/web/bin/MotifView/motif_hash_dump #stuff for Mauve and whole genome alignments MAUVE /opt/coge/web/bin/GenomeAlign/progressiveMauve-muscleMatrix COGE_MAUVE /opt/coge/web/bin/GenomeAlign/mauve_alignment.pl MAUVE_MATRIX /opt/coge/web/data/blast/matrix/nt/Mauve-Matrix-GenomeAlign #newicktops is part of njplot package NEWICKTOPS /usr/bin/newicktops #convert is from ImageMagick CONVERT /usr/bin/convert CUTADAPT /usr/local/bin/cutadapt GSNAP /usr/local/bin/gsnap CUFFLINKS /usr/local/bin/cufflinks PARSE_CUFFLINKS /opt/coge/scripts/parse_cufflinks.py GMAP_BUILD /usr/local/bin/gmap_build BOWTIE_BUILD /usr/local/bin/bowtie2-build TOPHAT /usr/local/bin/tophat #THIRD PARTY URLS GENFAMURL http://dev.gohelle.cirad.fr/genfam/?q=content/upload GRIMMURL http://grimm.ucsd.edu/cgi-bin/grimm.cgi#report QTELLER_URL http://geco.iplantc.org/qTeller
Install Perl Modules
- Install cpanminus
sudo cpan install App::cpanminus
- Install third-party modules required by CoGe
cat modules.txt | xargs sudo cpanm
- Install CoGe-specific modules
./make_perl.sh
- After installing modules, reset the Apache webserver
sudo service apache2 restart
Install Python Modules
sudo pip install pyzmq
Install Javascript dependencies
- Install javascript dependencies
sudo ln -s /usr/bin/nodejs /usr/bin/node sudo npm install -g bower bower install
Install Third-Party Bioinformatics Tools
For each path in coge.conf that starts with /usr/local/bin, download these programs and follow the installation instructions on their respective websites.
Most programs can be installed with the following commands (but check the documentation for each program):
./configure --prefix=/usr/local/ make sudo make install
- GSNAP/GMAP: http://research-pub.gene.com/gmap/
- FastBit: https://sdm.lbl.gov/fastbit/
- Clustalw: http://www.clustal.org/clustal2/
- Bowtie: http://bowtie-bio.sourceforge.net/bowtie2/index.shtml
- TopHat: http://ccb.jhu.edu/software/tophat/index.shtml
- HISAT2: https://ccb.jhu.edu/software/hisat2/index.shtml
- Cufflinks: http://cole-trapnell-lab.github.io/cufflinks/
- Nwalign: https://pypi.python.org/pypi/nwalign/?
- Cutadapt: http://cutadapt.readthedocs.io/en/stable/installation.html ... sudo su ; sudo pip install cutadapt
- TrimGalore: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- Lastz: download the tarball http://www.bx.psu.edu/~rsharris/lastz/ then edit the src/Makefile and remove the word -Werror from line 31. Then run make and make install.
- Last aligner (v731 or greater is required): http://last.cbrc.jp/
- VCFTools: https://github.com/vcftools/vcftools
- EMBOSS (sizeseq program): http://emboss.sourceforge.net/ ... Run "sudo ldconfig" after "make install"
- iCommands: https://github.com/irods/irods-legacy (OPTIONAL: only required if CyVerse authentication and Data Store services are available; note that the legacy version is required, not the latest)
- Bismark: http://www.bioinformatics.babraham.ac.uk/projects/bismark/
- BWAmeth:
- Homer: http://homer.salk.edu/homer/ ... sudo perl ./configureHomer.pl -install homer
- Blat (required by Homer): https://genome.ucsc.edu/FAQ/FAQblat.html
Install Third-Party Fonts
Download from here: https://www.microsoft.com/typography/fonts/font.aspx?FMID=1705
And copy to /usr/local/fonts/arial.ttf (or whatever path you set in the coge.conf config file under FONT)
Install blast matrices
cd /storage/coge/data/blast git clone https://github.com/LyonsLab/blast-matrix.git mv blast-matrix matrix
Install JBrowse
Copy from existing CoGe installation if one exists. Otherwise, download and install the JBrowse package from http://jbrowse.org/install/
unzip JBrowse-1.11.4-dev.zip mv JBrowse-1.11.4 /coge/web/js/jbrowse
Install CCTools
- Download a stable release that is 4.3 or greater from http://ccl.cse.nd.edu/software/downloadfiles.php
- Extract the file (this example is using version 4.3 which may differ from the version downloaded)
tar xzvf cctools-4.3.0-source.tar.gz
- Compile and install
cd cctools-4.3.0-source ./configure --prefix /usr/local make sudo make install
- Add the following upstart scripts for the work_queue_pool and catalog_server to /etc/init
By default the pool directory for work_queue will be in /storage/work_queue adjust the directory as needed.
# /etc/init/.conf
description "The cctools work queue pool"
author "Evan Briones"
start on (local-filesystems and net-device-up IFACE=eth0)
stop on shutdown
respawn limit 30 60
pre-start script
POOL_DIR=/storage/work_queue
LOG_FILE=$POOL_DIR/logs/work_queue_pool.log
# Add the pool directory and set ownership
if ! [ -d "$WORK_DIR" ]; then
mkdir -p $POOL_DIR/workers
mkdir -p $POOL_DIR/logs
chown -R www-data:www-data $POOL_DIR
fi
# Remove the pidfile if it exists
rm -f $POOL_DIR/work_queue_pool.pid
# Archive old log and timestamp the value
if [ -f "$LOG_FILE" ]; then
TIMESTAMP=$(date +"%Y-%m-%d.%H.%m.%S")
mv -f $LOG_FILE "$LOGFILE.$TIMESTAMP"
fi
end script
script
POOL_DIR=/storage/work_queue
LOG_FILE=$POOL_DIR/logs/work_queue_pool.log
#CONFIG=/etc/yerba/work_queue_pool.conf
#WORK_QUEUE_POOL=$(which work_queue_pool)
WORK_QUEUE_FACTORY=$(which work_queue_factory)
export CATALOG_HOST=localhost
export CATALOG_PORT=1024
exec start-stop-daemon -c www-data -g www-data -d $POOL_DIR --start \
-p $POOL_DIR/work_queue_pool.pid --exec $WORK_QUEUE_FACTORY \
-- -T local -M coge-main -d all -o $LOG_FILE -w 10 \
-S $POOL_DIR -E "--workdir=$POOL_DIR/workers"
end script
# /etc/init/.conf
description "The cctools catalog server"
author "Evan Briones"
start on (local-filesystems and net-device-up IFACE=eth0)
stop on shutdown
respawn limit 30 60
script
exec catalog_server -p 1024 -l 100 -T 3
end script
- Start the catalog server and work_queue_pool
sudo start work_queue_pool sudo start catalog_server
Install the Job Engine (Yerba)
Download and install the latest Yerba package from https://github.com/LyonsLab/Yerba/archive/v0.3.4.tar.gz
For more specific details on Yerba visit https://github.com/LyonsLab/Yerba/
The default installation path for Yerba will be in /opt/Yerba. If another path is chosen update the configuration files to match.
- Copy and the configuration file to /etc/yerba/yerba.cfg
[DEFAULT] debug = True access-log = /opt/Yerba/log/access.log yerba-log = /opt/Yerba/log/yerbad.log [yerba-log] logging = /etc/yerba/logging.conf [access-log] logging = /etc/yerba/access.conf [yerba] port = 5151 level = DEBUG [workqueue] catalog_server = localhost catalog_port = 1024 project = coge-main log = /var/log/workqueue.log port = -1 password = /etc/yerba/workqueue_pass debug = True [db] path = /opt/Yerba/workflows.db start_index = 100
- Copy the upstart file to /etc/upstart/yerba.conf
# /etc/init/yerba.conf
description "Yerba server daemon"
author "Evan Briones"
start on (local-filesystems and net-device-up IFACE=eth0)
stop on shutdown
respawn
pre-start script
LOG_DIR=/opt/Yerba/log
LOG_FILE=$LOG_DIR/debug.log
[ -d "$LOG_DIR" ] || mkdir -m777 -p $LOG_DIR
# [ -f "$LOG_FILE" ] || rm -f $LOG_FILE
end script
script
export YERBA_ROOT=/opt/Yerba
export PYTHONPATH="/usr/local/lib/python2.7/site-packages:$YERBA_ROOT"
exec start-stop-daemon -c www-data -g www-data --start \
--iosched real-time --nicelevel -19 \
--exec $YERBA_ROOT/bin/yerbad -- >> $YERBA_ROOT/log/debug.log 2>&1
end script
post-start script
echo Restart on: `hostname -A` | mail -s "UPSTART: Yerba was started" coge.genome@gmail.com
end script
- Initialize and start the job engine
/opt/Yerba/bin/yerbad --setup sudo chown www-data:www-data /opt/Yerba/workflows.db sudo start yerba
Troubleshooting
Visualization in GEvo does not work
This relies on a system known as Gobe. Check the following things:
- Apache configuration for gobe
- Check to see if paths hard-coded into gobe/flash/service.wsgi need to be updated
- NOTE: Not sure if this is required
Working on an Atmosphere Virtual Machine
Click here for instructions on dealing with issues that occur specifically with Atmosphere Virtual machines.