Variant data: Difference between revisions
Jump to navigation
Jump to search
Created page with 'EPIC-CoGe's genome browser supports the visualization of variant data on any genome in CoGe. You can load and visualize variant data by: # Loading a VCF file for a genome usin...' |
|||
| Line 15: | Line 15: | ||
====Text describing the variant is seen when density of variants is sufficiently low.==== | ====Text describing the variant is seen when density of variants is sufficiently low.==== | ||
[[ | [[File:Screen_Shot_2014-10-13_at_10.24.09_AM.png]] | ||
Revision as of 17:28, 13 October 2014
EPIC-CoGe's genome browser supports the visualization of variant data on any genome in CoGe.
You can load and visualize variant data by:
- Loading a VCF file for a genome using LoadExperiment
- Note: One loaded file in coge is called an experiment
- Using GenomeView to browse the genome
- Select the appropriate experiment for visualization
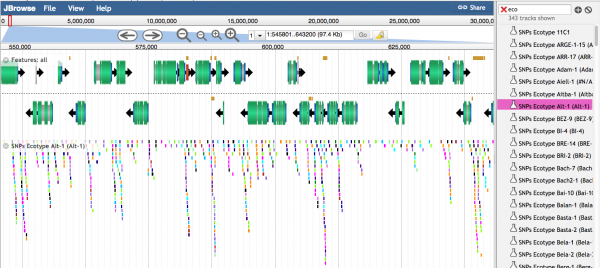
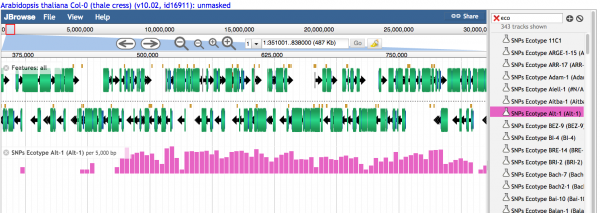
Visualizing Variant Data
Histograms showing the relative number of variants are visualized when the density is high
Individual variants are seen when the density of variants is lower. Different colors represent different types of SNPs