Sequenced plant genomes
After an 18 month hiatus, we are attempting to bring the CoGePedia list of plant genome sequences back up to date. If you know of publically avaliable plant genome sequences not listed here, please let us know!
This site attempts to track all plant genomes with published sequences, and at least some of the genomes currently in the process of being sequenced. Genomes are divided into four states:
- Published: A complete genome sequence is available, and anyone can publish papers on it without restriction.
- Unpublished: The complete sequence (or a substantially complete sequence) is available, but whole genome analysis cannot be published until the group that sequenced the genome publishes their own paper describing it. These restrictions are outlines by the Fort Lauderdale Convention.
- Incomplete: A partial assembly is available, but sequencing and/or assembly and/or gene annotation is ongoing.
- Unreleased: Genome sequencing has at least begun, but no data has been made publicly available.
Questions? Comments? Have we missed a published genome sequence? Get in touch and let me know!
For a table of sequenced plant genomes with additional statistics and information: Plant Genome Statistics
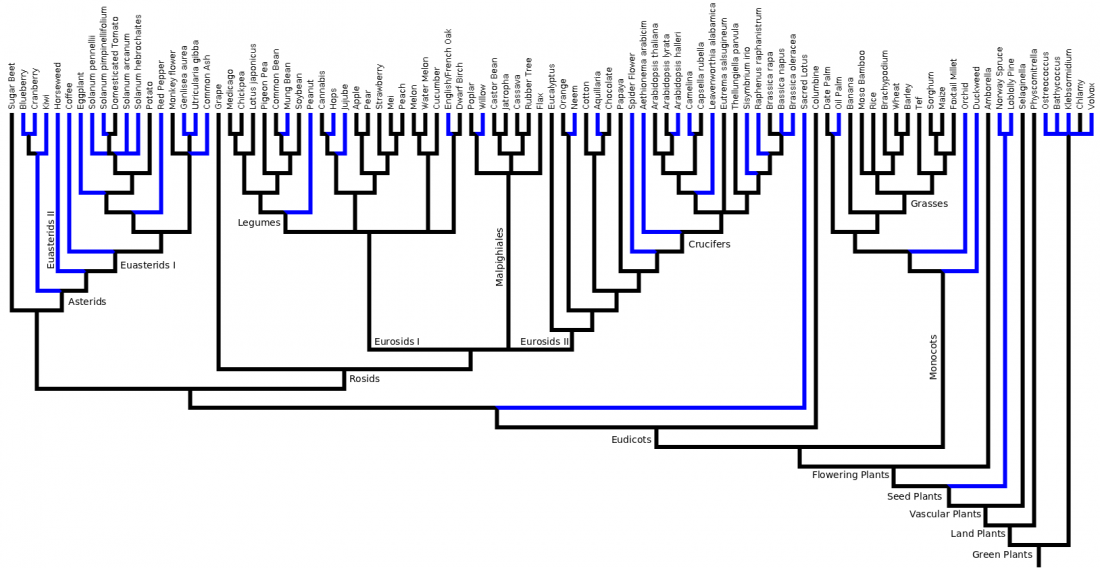
Phylogenetic Tree
Tree up to date as of Fall 2014
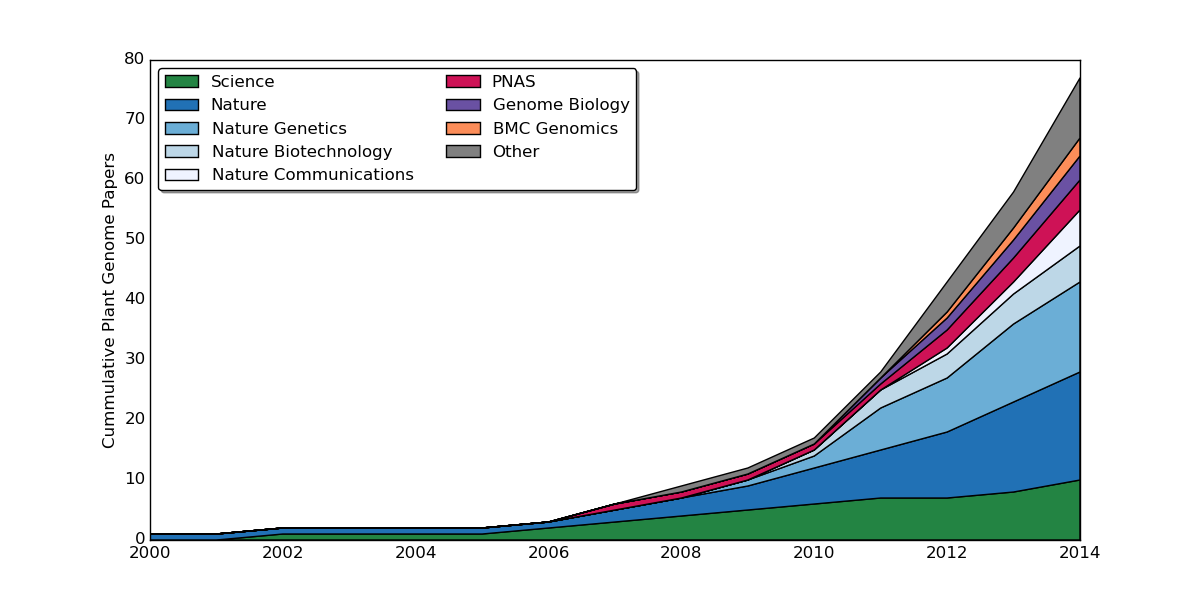
Rate of Publication
Graph up to date as of September 19th 2011
A detailed table of plant genome publications.
Amborella
Amborella (Amborella trichopoda)) is believed to represent the earliest diverging lineage of flowering plants (angiosperms) still alive today. While that doesn't mean it represents the ancestral state of flowering plants, comparing amborella to the major flowering plant lineages -- the eudicots, monocots, and magnoliids (the last of which still doesn't have even a single published genome, someone please get on that!) -- can tell us a lot about what that common ancestor. As the species is found only in New Caledonia and isn't exactly common even there, we are very fortunate that this sole representative species has survived to the present day.
The first draft of the Amborella trichopoda genome was released at the twentieth Plant and Animal Genome Conference in January of 2012. It is composed of 5,745 scaffolds covering 706 megabases out of an estimated total genome size of ~870 megabases. The genome is currently covered by Fort Lauderdale restrictions, but is available for download from the Amborella Sequencing project's website.
The Amborella Genome and the Evolution of Flowering Plants
Eudicots
The eudicots are the largest group of flowering plants on the planet.
Columbine
(genome available but unpublished)

Columbine (Aquilegia sp.) comes from a group of eudicots, the Ranunculales, whose ancestors split from the ancestors of the major eudicot groups (like rosids and asterids) a long, long, time ago (somewhere in the neighborhood of 115-130 million years ago). Comparing the columbine genome sequence with other eudicot genomes should be very interesting for several groups of plant biologists (comparative genomicists and evolutionary biologists in particular).
The columbine genome was sequenced to 8-fold coverage by JGI and a pre-publication release of the genome is available for download from phytozome (account creation/login required). The current assembly is only to the scaffold level (no pseudomolecules) and consists of 302 megabases of sequence spread over 971 scaffolds. Current gene annotations identify 25,784 genes identified by a mixture of EST sequencing and homology to other sequenced genomes. You can view in CoGe with GenomeView
As with all sequenced angiosperm genomes, columbine has an ancient whole genome duplication. However, is this the paleohexaploidy event shared among the rosids and asterids? Columbine's whole genome duplication
Sacred Lotus
Nelumbo nucifera
"Sacred lotus is a basal eudicot with agricultural, medicinal, cultural and religious importance. It was domesticated in Asia about 7,000 years ago, and cultivated for its rhizomes and seeds as a food crop. It is particularly noted for its 1,300-year seed longevity and exceptional water repellency, known as the lotus effect. The latter property is due to the nanoscopic closely packed protuberances of its self-cleaning leaf surface, which have been adapted for the manufacture of a self-cleaning industrial paint, Lotusan. The genome of the China Antique variety of the sacred lotus was sequenced with Illumina and 454 technologies, at respective depths of 101× and 5.2×. The final assembly has a contig N50 of 38.8 kbp and a scaffold N50 of 3.4 Mbp, and covers 86.5% of the estimated 929 Mbp total genome size. The genome notably lacks the paleo-triplication observed in other eudicots, but reveals a lineage-specific duplication. The genome has evidence of slow evolution, with a 30% slower nucleotide mutation rate than observed in grape. Comparisons of the available sequenced genomes suggest a minimum gene set for vascular plants of 4,223 genes. Strikingly, the sacred lotus has 16 COG2132 multi-copper oxidase family proteins with root-specific expression; these are involved in root meristem phosphate starvation, reflecting adaptation to limited nutrient availability in an aquatic environment. The slow nucleotide substitution rate makes the sacred lotus a better resource than the current standard, grape, for reconstructing the pan-eudicot genome, and should therefore accelerate comparative analysis between eudicots and monocots."
http://genomebiology.com/2013/14/5/R41
Sugar beet
The sugar beet -- a cultivar of the common beet (Beta vulgaris) -- accounts for ~20% of sugar production worldwide and is a favored crop in countries too cold to support a local sugar cane industry including Russia, much of the EU, and most of America. Sugar beets are a relatively recent agricultural innovation with selective breeding of beets for high sugar content only starting in 1784 and production not being adopted on a wide scale until the Napoleonic wars, during which large parts of Europe were essentially cut off from trade with the Caribbean, until then Europe's primary source of sugar from sugar cane.
Beets belong to the Caryophyllales an order of flowering plants which also includes the true cacti and many carnivorous plants. The Caryophyllales are currently believed to be more closely related to the Asterids than the Rosids, but are not included within either group.
The beet genome is currently at version 0.9 and encompasses 590 MB of sequence data split across 82,305 scaffolds and contigs. The genome is based upon a doubled haploid line called KWS2320. Version 1.0 is expected to include made improvements from correction of homopolymer errors introduced by next generation sequencing as well as integrating contigs using a genetic map.
For more information and to download the genome, visit the sugar beet genome sequencing group's website.
Asterids
The asterids are a group of plants within the eudicots that include species like the solanacious vegetables (Tobacco, Tomato, Potato, Peppers and Eggplant) and the sunflowers.
Domesticated Tomato
The tomato (Solanum lycopersicum) genome paper was published in Nature in May 2012. is not yet complete. The version of the genome currently loaded into CoGe is assembled into pseudomolecules[1]. The most recent assembly is 2.40 which includes 781 megabases of total sequence (out of an estimated genome size of ~900 megabases). Read more about the tomato genome project here
The Genome Paper The Tomato Genome Consortium (2012) "The tomato genome sequence provides insights into fleshy fruit evolution." Nature doi: [10.1038/nature11119]
Wild Tomatos
Solanum pennellii
"Solanum pennellii is a wild tomato species endemic to Andean regions in South America, where it has evolved to thrive in arid habitats. Because of its extreme stress tolerance and unusual morphology, it is an important donor of germplasm for the cultivated tomato Solanum lycopersicum. Introgression lines (ILs) in which large genomic regions of S. lycopersicum are replaced with the corresponding segments from S. pennellii can show remarkably superior agronomic performance. Here we describe a high-quality genome assembly of the parents of the IL population. By anchoring the S. pennellii genome to the genetic map, we define candidate genes for stress tolerance and provide evidence that transposable elements had a role in the evolution of these traits. Our work paves a path toward further tomato improvement and for deciphering the mechanisms underlying the myriad other agronomic traits that can be improved with S. pennellii germplasm."
Nature Genetics 2014 http://www.ncbi.nlm.nih.gov/pubmed/25064008
Solanum pimpinellifolium
This is a pre-publication genome
"Solanum pimpinellifolium is the wild tomato that is most closely related to the domesticated Solanum lycopersicum. It was sequenced by a group of scientists at the Cold Spring Harbor Laboratory."
http://solgenomics.net/organism/Solanum_pimpinellifolium/genome
Solanum arcanum
A perennial wild species related to tomato natuve to northern Peru.
Genome assembly published: http://onlinelibrary.wiley.com/doi/10.1111/tpj.12616/abstract
Solanum habrochaites
"Solanum habrochaites is a wild tomato species that is found on the Western slopes of the Andes from Central Ecuador to Central Peru. It has been used in a numerous genomic studies, such as identification of QTLs for traits of agronomic importance (1) or analysis of a set of 99 near isogenic lines (NILs) and backcross recombinant inbred lines (BCRILs) that cover more than 85% of Solanum habrochaites genome (2). It has also been used in biochemical analysis of fruit sucrose accumulation (3)." [From Solgenomics.net http://solgenomics.net/organism/938/view]
Genome assembly published: http://onlinelibrary.wiley.com/doi/10.1111/tpj.12616/abstract
Potato
Potatoes are arguably the second most important non-grass crop grown around the world. Both breeding and genomic analysis of the potato have been hampered by the fact that most cultivated potatoes are recent tetraploids. The genome of potato was published by an international consortium with corresponding authors hailing from the United States, China, and the Netherlands in 2011. It is the first publicly available genome from within the asterid clade. To avoid the complexities introduced by tetraploidy, the genome consortium focused on a diploid potato variety and used doubled-monoploid technology to create an "instantly inbred line." This assembled genome was used as a base to analyze further data generated from a hetrozygous line where a great deal of presence absence variation was detected. The potato lineage has experienced one additional tetraploidy since the ancient hexaploidy shared by the asterids and rosids.
The current genome assembly contains an estimated 86% of the total potato genome, and 74% of the total potato genome has been assembled into 12 pseudomolecules using genetic and physical maps. A total of 39,031 protein coding genes were annotated in the current assembly.
The Genome Paper: The Potato Genome Sequencing Consortium (2011). Genome sequence and analysis of the tuber crop potato. Nature, 475: 189–195 DOI 10.1038/nature10158
Eggplant (Solanum melongena L.)
"Unlike other important Solanaceae crops such as tomato, potato, chili pepper, and tobacco, all of which originated in South America and are cultivated worldwide, eggplant (Solanum melongena L.) is indigenous to the Old World and in this respect it is phylogenetically unique. To broaden our knowledge of the genomic nature of solanaceous plants further, we dissected the eggplant genome and built a draft genome dataset with 33,873 scaffolds termed SME_r2.5.1 that covers 833.1 Mb, ca. 74% of the eggplant genome. Approximately 90% of the gene space was estimated to be covered by SME_r2.5.1 and 85,446 genes were predicted in the genome. Clustering analysis of the predicted genes of eggplant along with the genes of three other solanaceous plants as well as Arabidopsis thaliana revealed that, of the 35,000 clusters generated, 4,018 were exclusively composed of eggplant genes that would perhaps confer eggplant-specific traits. Between eggplant and tomato, 16,573 pairs of genes were deduced to be orthologous, and 9,489 eggplant scaffolds could be mapped onto the tomato genome. Furthermore, 56 conserved synteny blocks were identified between the two species. The detailed comparative analysis of the eggplant and tomato genomes will facilitate our understanding of the genomic architecture of solanaceous plants, which will contribute to cultivation and further utilization of these crops." --DNA Research Abstract
Monkey Flower
(genome incomplete)
The monkey flower (Mimulus guttatus) genome is not yet complete. The version of the genome currently loaded into CoGe is not assembled into pseudomolecules[1] but does contain genome models Read more about the monkey flower genome on phytozome (account creation/login required) or see the current assembly in GenomeView here.
Blueberry
"Background: Blueberries are a rich source of antioxidants and other beneficial compounds that can protect against disease. Identifying genes involved in synthesis of bioactive compounds could enable breeding berry varieties with enhanced health benefits. Results: Toward this end, we annotated a draft blueberry genome assembly using RNA-Seq data from five stages of berry fruit development and ripening. Genome-guided assembly of RNA-Seq read alignments combined with output from ab initio gene finders produced around 60,000 gene models, of which more than half were similar to proteins from other species, typically the grape Vitis vinifera. Comparison of gene models to the PlantCyc database of metabolic pathway enzymes identified candidate genes involved in synthesis of bioactive compounds, including bixin, an apocarotenoid with potential disease-fighting properties, and defense-related cyanogenic glycosides, which are toxic. Cyanogenic glycoside (CG) biosynthetic enzymes were highly expressed in green fruit, and a candidate CG detoxification enzyme was up regulated during fruit ripening. Candidate genes for ethylene, anthocyanin, and 400 other biosynthetic pathways were also identified. RNA-Seq expression profiling showed that blueberry growth, maturation, and ripening involve dynamic gene expression changes, including coordinated up and down regulation of metabolic pathway enzymes, cell growth-related genes, and putative transcriptional regulators. Analysis of RNA-seq alignments also identified developmentally regulated alternative splicing, promoter use, and 3' end formation. Conclusions: We report genome sequence, gene models, functional annotations, and RNA-Seq expression data which provide an important new resource enabling high throughput studies in blueberry. RNA-Seq data are freely available for visualization in Integrated Genome Browser, and analysis code is available from the git repository at http://bitbucket.org/lorainelab/blueberrygenome."
http://biorxiv.org/content/early/2014/10/08/010116
Coffee
"Coffee is a valuable beverage crop due to its characteristic flavor, aroma, and the stimulating effects of caffeine. We generated a high-quality draft genome of the species Coffea canephora, which displays a conserved chromosomal gene order among asterid angiosperms. Although it shows no sign of the whole-genome triplication identified in Solanaceae species such as tomato, the genome includes several species-specific gene family expansions, among them N-methyltransferases (NMTs) involved in caffeine production, defense-related genes, and alkaloid and flavonoid enzymes involved in secondary compound synthesis. Comparative analyses of caffeine NMTs demonstrate that these genes expanded through sequential tandem duplications independently of genes from cacao and tea, suggesting that caffeine in eudicots is of polyphyletic origin."
[The coffee genome provides insight into the convergent evolution of caffeine biosynthesis http://www.sciencemag.org/content/345/6201/1181.full]
Utricularia gibba
The smallest flowering plant genome sequenced to date. (Statement accurate as of Fall 2014)
"It has been argued that the evolution of plant genome size is principally unidirectional and increasing owing to the varied action of whole-genome duplications (WGDs) and mobile element proliferation1. However, extreme genome size reductions have been reported in the angiosperm family tree. Here we report the sequence of the 82-megabase genome of the carnivorous bladderwort plant Utricularia gibba. Despite its tiny size, the U. gibba genome accommodates a typical number of genes for a plant, with the main difference from other plant genomes arising from a drastic reduction in non-genic DNA. Unexpectedly, we identified at least three rounds of WGD in U. gibba since common ancestry with tomato (Solanum) and grape (Vitis). The compressed architecture of the U. gibba genome indicates that a small fraction of intergenic DNA, with few or no active retrotransposons, is sufficient to regulate and integrate all the processes required for the development and reproduction of a complex organism."
Architecture and evolution of a minute plant genome Nature 2013
Rosids
Grape

The genome sequence of the wine grape (Vitis vinifera) was published by a group of French and Italian researchers in 2007. The variety of grape sequenced was the Pinot Noir.
Grape diverged early from the two main groups of species in the rosids (eurosids I and eurosids II) and has not experienced any whole genome duplications since that divergence making it an important outgroup for comparisons to other rosid species as well as providing a great resource for studying the ancient hexaploidy that preceeding the radiation of rosid species (and possibly the radiation of eudicot species).
The version of the grape genome in CoGe contains ~500 megabases of sequence and 26346 annotated genes spread across 19 chromosomes.
The genome paper:
Jaillon, O., Aury, J., Noel, B., Policriti, A., Clepet, C., Casagrande, A., Choisne, N., Aubourg, S., Vitulo, N., Jubin, C., Vezzi, A., Legeai, F., Hugueney, P., Dasilva, C., Horner, D., Mica, E., Jublot, D., Poulain, J., Bruyère, C., Billault, A., Segurens, B., Gouyvenoux, M., Ugarte, E., Cattonaro, F., Anthouard, V., Vico, V., Del Fabbro, C., Alaux, M., Di Gaspero, G., Dumas, V., Felice, N., Paillard, S., Juman, I., Moroldo, M., Scalabrin, S., Canaguier, A., Le Clainche, I., Malacrida, G., Durand, E., Pesole, G., Laucou, V., Chatelet, P., Merdinoglu, D., Delledonne, M., Pezzotti, M., Lecharny, A., Scarpelli, C., Artiguenave, F., Pè, M., Valle, G., Morgante, M., Caboche, M., Adam-Blondon, A., Weissenbach, J., Quétier, F., & Wincker, P. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla Nature, 449 (7161), 463-467 DOI: 10.1038/nature06148
Rose Gum Tree/Eucalyptus
One of several species of tree referred to by the common name "Eucalyptus", the rose gum tree (Eucalyptus grandis) is native to Australia, but is considered a candidate for biofuel production in the US. The rose gum tree is a basal rosid, like grape, so in addition to the value of this genome sequence for biofuel breeding purposes, this genome serves as a valuable outgroup for the core rosids (listed as Eurosids 1 and Eurosids 2 on this site).
The rose gum genome was sequenced to 8x coverage by the Joint Genome Institute and is assembled into 11 linkage/chromosome groups. The initial release of the genome includes 691 MB of sequence data, and 41,204 protein coding genes located on the putative chromosome assemblies. Read more/download the sequence from Phytozome (account creation/login required).
[The genome sequencing paper is here http://www.nature.com/nature/journal/v510/n7505/full/nature13308.html]
Malpighiales
Different people using different lines of evidence debate whether the Malpighiales belong to Eurosids I or Eurosids II. Until that uncertainly is resolved they are presented as a separate branch within the Rosids.
Poplar

The genome sequence of the black cottonwood tree (Populus trichocarpa) was published in 2006. The genome was originally sequenced to a coverage of 7.5x using Sanger sequencing. Poplar was the third plant genome to be published, and is now one of two published genomes of tree species (the other being papaya). Poplar contains a whole genome duplication that is not shared by any other plant species with a sequenced genome. The most recent version of the poplar genome in CoGe is v2 available on Phytozome (account creation/login required) which includes ~370 megabases of sequence and 41377 protein coding genes spread over 19 chromosomes.
The genome paper: Tuskan, G., DiFazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., Putnam, N., Ralph, S., Rombauts, S., Salamov, A., Schein, J., Sterck, L., Aerts, A., Bhalerao, R., Bhalerao, R., Blaudez, D., Boerjan, W., Brun, A., Brunner, A., Busov, V., Campbell, M., Carlson, J., Chalot, M., Chapman, J., Chen, G., Cooper, D., Coutinho, P., Couturier, J., Covert, S., Cronk, Q., Cunningham, R., Davis, J., Degroeve, S., Dejardin, A., dePamphilis, C., Detter, J., Dirks, B., Dubchak, I., Duplessis, S., Ehlting, J., Ellis, B., Gendler, K., Goodstein, D., Gribskov, M., Grimwood, J., Groover, A., Gunter, L., Hamberger, B., Heinze, B., Helariutta, Y., Henrissat, B., Holligan, D., Holt, R., Huang, W., Islam-Faridi, N., Jones, S., Jones-Rhoades, M., Jorgensen, R., Joshi, C., Kangasjarvi, J., Karlsson, J., Kelleher, C., Kirkpatrick, R., Kirst, M., Kohler, A., Kalluri, U., Larimer, F., Leebens-Mack, J., Leple, J., Locascio, P., Lou, Y., Lucas, S., Martin, F., Montanini, B., Napoli, C., Nelson, D., Nelson, C., Nieminen, K., Nilsson, O., Pereda, V., Peter, G., Philippe, R., Pilate, G., Poliakov, A., Razumovskaya, J., Richardson, P., Rinaldi, C., Ritland, K., Rouze, P., Ryaboy, D., Schmutz, J., Schrader, J., Segerman, B., Shin, H., Siddiqui, A., Sterky, F., Terry, A., Tsai, C., Uberbacher, E., Unneberg, P., Vahala, J., Wall, K., Wessler, S., Yang, G., Yin, T., Douglas, C., Marra, M., Sandberg, G., Van de Peer, Y., & Rokhsar, D. (2006). The Genome of Black Cottonwood, Populus trichocarpa (Torr. & Gray) Science, 313 (5793), 1596-1604 DOI: 10.1126/science.1128691
Shrub Willow
Salix purpurea Unpublished but available under Ft. Lauderdale
"This is an early release v1.0 of the genome sequence of Salix purpurea female clone 94006. Salix purpurea is a diploid shrub that is native to Europe and is naturalized in North America. Clone 94006 was collected from the banks of a river just north of Syracuse, NY. Salix purpurea and interspecific hybrids are being developed as a clonally propagated bioenergy crop that is managed in short-rotation coppice systems that are typically harvested every three years with vigorous resprouting after each harvest. Salix purpurea is a close relative of the DOE flagship model tree species, Populus trichocarpa, in the family Salicaceae."
From Phytozome at JGI (where you can also download the genome after creating a JGI account and logging in).
Flax
Flax (Linum usitatissimum) is an ancient fiber crop grown to produce linen and is also used as an oilseed crop to produce linseed oil (also called, you guessed it "flaxseed oil"). Flax has a small total genome size (estimated to be ~350 megabases) and the current assembly v1.0 was produced entirely by Illumina sequencing. This early assembly consists of a huge number of scaffolds (>88,000) however 290 megabases of the flax genome is present in only 664 scaffolds, a far more manageable number. The flax genome project is a collaboration between BGI and a group of canadian researchers. Download through phytozome (account creation/login required).
The Genome Paper: Wang Z. (2012) "The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads." The Plant Journal DOI: 10.1111/j.1365-313X.2012.05093.x
Castor Bean
The castor bean (Ricinus communis) is an oilseed plant that is the source of castor oil and the deadly poison ricin. The castor bean should not to be confused with the common bean (Phaseolus vulgaris) which is in the Joint Genome Institute sequencing pipeline.
The published castor bean genome is based on a 4.6x coverage of the genome using solexa sequencing.
The current release consists of 31,237 gene models spread across 25,800 scaffolds.
The entire genome is estimated to be ~320 megabases in size and contains 10 chromosomes.
Genome Paper Agnes P Chan et al., “Draft genome sequence of the oilseed species Ricinus communis,” Nature Biotechiology, DOI 10.1038/nbt.1674
The website of the castor bean sequencing group.
Cassava
Cassava (Manihot esculenta) is the most important crop that most people in America and Europe have never heard of (except perhaps in the form of tapioca). Originally domesticated in South America, cassava is now an important food source in Southeast Asia and Africa. The current draft genome is made available through phytozome (account creation/login required) and consists of 416 megabases of sequence spread over 11,243 contigs. This is only a little over 50% of the estimated total size of the cassava genome, but the people involved in the sequencing and assembly believe it represents the majority of the non-repetitive genome. The current release also includes 47,164 predicted genes.
The Genome Paper: Prochnik S et al (2012) "The Cassava Genome: Current Progress, Future Directions." DOI: 10.1007/12042-011-9088-z <-- This really is the paper Phytozome says to cite when you use the cassava genome.
Rubber Tree
While other plants also produce natural rubber the vast majority of what is produced commercially comes from this species (Hevea brasiliensis). While the current genome assembly covers only half the estimated genome size of the rubber tree (1.1 GB assembled from a 2.15 GB genome) and is highly fragmented (N50 = ~3,000), the authors present evidence that their assembly captures the majority of the rubber genome gene space.
The current genome annotation contains nearly 70,000 putative genes based on protein alignments from related species, RNA-seq, and de novo gene prediction software packages. The rubber tree genome is made up of 18 chromosomes.
The Genome Papper: Yamin Abdul Rahman, A et al (2013) "Draft genome sequence of the rubber tree Hevea brasiliensis." BMC Genomics doi: 10.1186/1471-2164-14-75
Right now it appears the only place to grab the rubber genome assembly from in NCBI.
Eurosids 1
Dwarf Birch
The dwarf birch ('Betula nana'). Add more details here.
The Genome Paper: Wang N et al (2012) "Genome sequence of dwarf birch (Betula nana) and cross-species RAD markers." Molecular Ecology DOI: 10.1111/mec.12131
Cucurbits
Cucumber

The genome sequence of cucumber (Cucumis sativus) was published in late 2009. The genome of the inbred line "'Chinese long' 9930" was sequenced using a combination of Illumina short read sequencing (68.3x coverage) and Sanger sequencing (3.9x coverage). The complete published genome consists of 243.5 megabases of sequence and 26,682 protein coding genes, 72.8% of which can been anchered to the seven cucumber chromosomes, with the rest unanchored.
The publication also included a genetic map with a total length of 581 cM based on 1,885 markers. Resources from this version of the cucumber genome are available at this site.
Independently a group of researchers in the US have released a draft of a cucumber genome sequence of the inbred line Gy14. This version of the genome was assembled from 454 sequencing reads and the current release consists of 203 megabases of sequence and a predicted 21491 protein coding genes spread over 4219 scaffolds. This version is available at Phytozome.
A third version of the cucumber genome, this one of the cultivar Borszczagowski line B10 was produced by a Polish research group and published in 2011 and the resulting data is available here.
The genome paper: Huang, S., Li, R., Zhang, Z., Li, L., Gu, X., Fan, W., Lucas, W., Wang, X., Xie, B., Ni, P., Ren, Y., Zhu, H., Li, J., Lin, K., Jin, W., Fei, Z., Li, G., Staub, J., Kilian, A., van der Vossen, E., Wu, Y., Guo, J., He, J., Jia, Z., Ren, Y., Tian, G., Lu, Y., Ruan, J., Qian, W., Wang, M., Huang, Q., Li, B., Xuan, Z., Cao, J., Asan, ., Wu, Z., Zhang, J., Cai, Q., Bai, Y., Zhao, B., Han, Y., Li, Y., Li, X., Wang, S., Shi, Q., Liu, S., Cho, W., Kim, J., Xu, Y., Heller-Uszynska, K., Miao, H., Cheng, Z., Zhang, S., Wu, J., Yang, Y., Kang, H., Li, M., Liang, H., Ren, X., Shi, Z., Wen, M., Jian, M., Yang, H., Zhang, G., Yang, Z., Chen, R., Liu, S., Li, J., Ma, L., Liu, H., Zhou, Y., Zhao, J., Fang, X., Li, G., Fang, L., Li, Y., Liu, D., Zheng, H., Zhang, Y., Qin, N., Li, Z., Yang, G., Yang, S., Bolund, L., Kristiansen, K., Zheng, H., Li, S., Zhang, X., Yang, H., Wang, J., Sun, R., Zhang, B., Jiang, S., Wang, J., Du, Y., & Li, S. (2009). The genome of the cucumber, Cucumis sativus L. Nature Genetics, 41(12), 1275-1281 DOI: 10.1038/ng.475
Melon
Melon (Cucumis melo is a close relative of cucumber (you can tell because they share the same genus name... or by remembering how similar the plants looked if you had a garden growing up). The sequenced melon genome is of a doubled haploid line called DHL92 and is estimated to cover 83% of the total genome (375 and 450 megabases respectively). The genome was sequenced primary with 454 reads (13.5x coverage) although Sanger sequencing of BAC ends were used to assist in the assembly, and Illumina reads were used to correct errors in homopolymer regions (a series of AAAA, TTTT, CCCC, or GGGG <-- 454 has trouble figuring out how many total copies of a nucleotide are present is run on sequences like these).
The genome was assembled into 12 pseodomolecules with the assistance of a genetic map. These twelve pseudomolecules contain 316 megabases of sequence (roughly 70% of the total estimated melon genome size).
The Genome Paper:
Garci-Mas J et al (2012) "The Genome of melon (Cucumis melo L.)" PNAS DOI: 10.1073/pnas.1205415109
Download sequence data:
Water Melon
The water melon genome (Citrullus lanatus) is another genome brought to us by the Beijing Genomics Institute. The 425 megabase genome was sequenced to a depth of >100x using Illumina short read sequencing. The resulting genome assembly included 353.5 megabases of sequence, 330 megabases of which were placed into 11 pseudomolecules representing each watermelon chromosome.
Download the watermelon genome here.
The Genome Paper: Guo et al (2012) "The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions" Nature Genetics DOI: 10.1038/ng.2470
Woodland Strawberry
The woodland strawberry (Fragaria vesca) is not the species that produces most of the strawberries you see on grocery store shelves today. Those are generally from the garden strawberry. However garden strawberries are octoploid, making sequencing their genome relatively difficult, while the woodland strawberry possesses a much more manageable diploid genome. For more on the story of how the woodland strawberry came to be sequenced, check out this fascinating story from one of the scientists behind the genome paper.
The published strawberry genome consists of 7 chromosomes/pseudomolecules.
The Genome Paper: Vladimir Shulaev et al., "The genome of woodland strawberry (Fragaria vesca)," Nature Genetics 43: 109-116. DOI: 10.1038/ng.740
Apple
The Apple (Malus x domestica) genome was published in late August of 2010. The total genome is estimated to be 742.3 MB large, spread over 17 chromosomes. The published genome includes 600 megabases of sequence assembled into 17 pseudomolecules and a number of smaller unanchored contigs. The apple genome contains 57,386 putative genes, a high number attributable, at least in part, to a whole genome duplication in the apple lineage which is dated to 30-65 million years ago. The apple genome is not yet loaded into CoGe, and does not yet appear to be available for download, however, there is an available genome browser.
Genome Paper
Riccardo Velasco et al., “The genome of the domesticated apple (Malus [times] domestica Borkh.),” Nature Genetics, DOI: 10.1038/ng.654
Pear
The Pear (Pyrus bretschneideri Rehd. cv. Dangshansuli) genome has been sequenced by chinese group based at Nanjing Agricultural University. The genome paper was not initially published, however it is possible to request a prepublication copy of the genome through the pear genome project website. The genome paper was released in November 2012.
While I haven't looked at the pear genome assembly myself (most of the stuff I'm interested in would count as reserved analyses) the description of the genome is very promising, as the researchers say they used a BAC-by-BAC approach to sequencing and also generated a dense genetic map covering all 17 pear chromosomes. Both these approaches will result in larger and more accurate genome assemblies than are possible when using short read sequencing technologies like Illumina and Ion torrent. (The downside is BAC-by-BAC sequencing takes longer and costs more, which is why it isn't as common anymore).
Wu et al (2012) "The genome of pear (Pyrus bretschneideri Rehd.)" Genome Research doi:10.1101/gr.144311.112
Cannabis
The genome of cannabis (Cannabis sativa) was published in Genome Biology in October 2011. The genome sequence was completed using a mixture 454 and Illumina sequencing, with mate pairs used to bridge gaps in the assembled regions. As a result, while only 534 megabases of the genome were assembled the genome spans >786 Mb of sequence (the extra 200 MB are NNNNNN's representing unassembled repeat sequences -- transposons -- of known length between sequenced regions of the genome). In addition to the genome itself, the same research group generated a great deal of tissue specific RNA-seq data from multiple cannabis cultivars.
Genome Paper: Harm van Bakel et al "The draft genome and transcriptome of Cannabis sativa." Genome Biology DOI: 10.1186/gb-2011-12-10-r102
Links
Peach

Peaches (Prunus persica) are stone fruits, meaning they're closely related to fruits such as plums, apricots, and cherries and nuts like almonds. The 1.0 version of the peach genome assembly was released by the International Peach Genome Initiative on April 1st, 2010. This version of the genome is already assembled into eight pseudomolecules covering the eight chromosomes of peach, as well as ~200 smaller unplaced contigs. The total released sequence is 227 megabases and includes 27,852 annotated genes. The genome was sequenced to 7.7x coverage using Sanger sequencing.
In March 2013 the paper describing the peach genome was published in Nature Genetics:
International Peach Genome Initiative 2013 "The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution" DOI: 10.1038/ng.2586
Chinese Plum/Mei
The Chinese plum (or Japanese Apricot, basically europeans weren't creative at all about coming up with english names for asian produce) is, like peach, a member of the genus Prunus. Specifically Prunus mume. Also like peach it has a ridiculously small genome (280 megabases) which has been well assembled into eight pseudomolecules using both genetic and optical mapping. The Chinese plum genome was sequenced to 100x coverage using Illumina sequencing.
The genome data and annotations are provided by this website: http://prunusmumegenome.bjfu.edu.cn/
Genome Paper:
Qixiang Zhang et al "The genome of Prunus mume." Genome Biology DOI: 10.1038/ncomms2290
Legumes
Legumes (the plant family Fabaceae) contained within the eurosid II clade. The family is perhaps best known for the fact that many of the species it contains form symbiotic relationships with nitrogen fixing bacteria. The bacteria are sheltered and feed within special nodules in the roots of these plants and in return the plant benefits from the bacteria's ability to convert the nitrogen in our atmosphere into bio-available forms (bioavailable nitrogen is often a limiting nutrient for other plant species).
Medicago
Medicago (Medicago truncatula) is small legume used as a model species for nodule formation and nitrogen fixing -- as is Lotus. The latest release of the medicago genome is Mt3.0 which includes 240 megabases of sequence associated with Medicago's eight chromosomes, plus 16.6 megabases of unanchored sequence. Read more at International Medicago Genome Annotation Group's webpage.
Genome paper: Young ND et al (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature DOI: 10.1038/nature10625
Chickpea
The Chickpea (Cicer arietinum) is widely grown around the world although the centers of production and consumption are the middle east and India. While chickpeas were the main ingredient of the delicious chana masala that sustained your humble author through many a late night hunched over his computer in grad school, in most western cuisine chickpeas will most often be encountered mashed up to make hummus. Depending the the grocery store canned chickpeas may also be labeled as garbanzo beans. But on to the genome!
The chickpea genome is derived from the accession CDC Frontier which is member of the kabuli subtype. Based on kmer abundance the authors estimate the total genome size to be ~740 megabases in size. Using a whole lot of Illumina data (>100-fold coverage of the genome after quality trimming) the authors were able to assemble 545 megabases into contigs. Using genetic maps and BAC end sequences the authors were able to place 345 megabases on sequence onto eight pseudomolecules.
Genome Paper: Varshney RK et al (2013) "Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement." Nature Biotechnology DOI:10.1038/nbt.2491
A second chickpea genome has now also been published. This project targeted the desi type chickpea and resulting in a 520 megabase genome assembly:
Genome Paper: Jain M et al (2013) "A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.)." The Plant Journal DOI: 10.1111/tpj.12173
Lotus japonicus
Lotus japonicus is a small legume used as a model for nodule formation and nitrogen fixation -- as is Medicago. The current release of the Lotus genome is v2.5 which includes 315 megabases of assembled sequence (an estimated 67% of the genome). In v2.5, 201 megabases of sequence have been assembled into six pseudomolecules corresponding to the six chromosomes of Lotus. Additional statistics and links to download the genome are proved by the Kazusa DNA Research Institute.
Genome paper: Sato S et al (2008) Genome Structure of the Legume, Lotus japonicus. DNA Research DOI: 10.1093/dnares/dsn008
Soybean

Soybeans (Glycine max) are an important crop species valued both as a source of protein and for their ability to fix nitrogen, which reduces the amount of fertilizer that needs to be applied to whatever crop is grown in the same field the following year.
The soybean genome was published in early 2010 and contained 950 megabases of sequence as well as a predicted 46,430 protein coding genes distributed over twenty chromosomes. The ancestors of soybean went through two whole genome duplications since the ancient hexaploidy as the base of the eudicot lineage with the older estimated to have occured 59 million years ago and the more recent estimated to have occured 13 million years ago.
The Genome Paper: Schmutz, J., Cannon, S., Schlueter, J., Ma, J., Mitros, T., Nelson, W., Hyten, D., Song, Q., Thelen, J., Cheng, J., Xu, D., Hellsten, U., May, G., Yu, Y., Sakurai, T., Umezawa, T., Bhattacharyya, M., Sandhu, D., Valliyodan, B., Lindquist, E., Peto, M., Grant, D., Shu, S., Goodstein, D., Barry, K., Futrell-Griggs, M., Abernathy, B., Du, J., Tian, Z., Zhu, L., Gill, N., Joshi, T., Libault, M., Sethuraman, A., Zhang, X., Shinozaki, K., Nguyen, H., Wing, R., Cregan, P., Specht, J., Grimwood, J., Rokhsar, D., Stacey, G., Shoemaker, R., & Jackson, S. (2010). Genome sequence of the palaeopolyploid soybean Nature, 463 (7278), 178-183 DOI: 10.1038/nature08670
- A wild relative of the domesticated soybean (Glycene soja) has been resequenced using Illumina technology and aligning reads to the existing Glycine max assembly. The paper describing this re-sequencing effort can be found here.
Pigeon Pea
Pigeon peas (Cajanus cajan) are grown in areas with low rainfall as an important source of protein for farmers and an important source of fixed nitrogen in the soil for whichever crop is grown the following year. They are consider an orphan crop (a species of great importance to feeding people around the world -- the main source of protein for 1 BILLION PEOPLE according the the genome paper -- but grown primarily by small farmers in developing countries, which means the species hasn't benefitted from the yield increases that can be produced by modern breeding practices).
The pigeon pea genome was published in Nature Biotechnology in November 2011. The genome was sequenced primarily with Illumina short reads, although assembly was assisted by a number of BAC send sequences produced using traditional Sanger-sequencing long reads. The assembly contains 606 megabases of sequence, a little under three quarters of the estimated total genome size of 833 megabases, and includes an estimated 48,680 genes. While the pidgeon pea genome is made up of 11 chromosomes, the current assembly consists of ~7,000 super scaffolds.
The Genome Paper: Varshney RK et al (2011) Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nature Biotechnology DOI: 10.1038/nbt.2022
Common bean

The common bean (Phaseolus vulgaris) is the single species into which beans of varieties ranging from "string" to "pinto" to "black" fall and the shapes and growth habits of common bean plants are at least as diverse as the varieties of beans they produce. Dried beans are an essential source of protein especially for grad students and others trying to feed themselves (or their families) on severely limited budgets. But what about the genome?
The current release of the common bean genome is 1.0 and was assembled from ~20x coverage of the genome using 454 reads (and a smaller number of paired end reads). This assembly included 521 megabases of assembled sequence and as been assembled to the scaffold level. By adding in BAC and fosmid end sequences as well as a genetic map with 7,015 markers (that's a lot!) version 1.0 was able to increase assembly from the scaffold to the pseudomolecule level. Read more and download the genome at phytozome (account creation/login required).
The common bean genome is published and now freely available:
A reference genome for common bean and genome wide analysis of dual domestications Published in Nature in 2014.
Peanut
Unpublished but available for download.
"Cultivated peanut, Arachis hypogaea, is an allotetraploid (2n=4x=40) that contains two complete genomes, labeled the A and B genomes. A. duranensis (2n=2x=20) has likely contributed the A genome, and A. ipaensis has likely contributed the B genome. It may be helpful to remember these two associations by using the mnemonic: "A" comes before "B" and "duranensis" comes before "ipaensis"." Currently the two diploid progenitors have both been sequenced and the genome assemblies may be downloaded from the link below (also the source of the quoted text above).
Eurosids 2
Cotton
The first (potentially of several) cotton species to have its genome sequenced is Gossypium raimonddi. G. raimonddi contributes the "D" genome to the allotetraploid cotton species (A + D genomes) G. hirsutum which provides the majority of worldwide cotton production. The genome of G. raimonddi was sequenced by JGI and is available from phytozome but has not yet been published.
The current genome assembly represents ~750 megabases of sequence and 98% of it is incorporated into 13 pseudomolecules and another 22 large unplaced scaffolds (> 50 kb).
A second cotton genome assembly was published by a group of Chinese scientists working with BGI in August 2012. As of this writing the BGI genome assembly doesn't appear to be available to download anywhere. The paper contains this link which leads to a "coming soon" webpage. Hopefully you have more luck when visiting this page in the future.
BGI version's stats: 775 MB of total assembled sequence, 570 MB incorporated into 13 pseudomolecules. 40,976 annotated genes, most of which were supported by RNA-seq data.
Genome Paper (BGI):
Wang K et al (2012) "The draft genome of a diploid cotton Gossypium raimondii" Nature Genetics doi:10.1038/ng.2371
Chocolate
The genome of the tree that gives us chocolate Theobroma cacao has been independently sequenced by two groups. One genome assembly, of the variety called Criollo from Belize has been in the Nature Genetics. A second assembly of a breed called Matina 1-6 has available from the Cacao genome database since before the publication of the Criollo genome sequence, but has not yet been published. Both assemblies are complete to the level of pseudomolecules.
Chocolate has not experienced any whole genome duplications since the ancient hexaploidy shared by all sequenced rosids.
Genome Paper (Criollo version):
Xavier Argout et al., "The genome of Theobroma cacao," Nature Genetics 43 (2): 101-108. DOI: 10.1038/ng.736
Neem
Neem (Azadirachta indica) is a relative of relative of mahogany found in India and neighboring countries. The tree has a wide range of uses ranging from edible flowers, providing an oil used in various soaps, and serving as an insect repellent. The genome of Neem is approximately 370 megabases in size. Sequence data for Neem can be downloaded here.
Genome Paper: Krishnan NM et al (2012) "A draft of the genome and four transcriptomes of a medicinal and pesticidal angiosperm Azadirachta indica" BMC Genomics DOI: 10.1186/1471-2164-13-464
Citrus fruits (genus Citrus)
Citrus fruits from lemons to oranges, grapefruits and pomelos belong to a singe genus. Many fruits we think of as separate species can breed with each other, making it difficult to properly define species barriers.
Sweet/Common Orange
The sweet orange (Citrus sinensis) was sequenced using a combination of Sanger (old fashion, expensive, but long and easy to assemble) and 454 (much cheaper, faster, and somewhat shorter) sequencing technology. The current release is only version 0.1 and the genome is still split into 12,574 scaffolds that cover a combined 319 megabases of the sweet orange genome. Unlike the clementine genome described below, the sweet orange genome project used DNA from a diploid individual, making the assembly of the genome somewhat more difficult as inconsistences between aligned sequences might simply be the result of variation between the two genome copies of that diploid individual. This version of the genome release includes 25,376 annotated protein coding genes. You can read more or download data here (account creation/login required).
Genome paper:
Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication in Nature Biotech 2014.
Clementine mandarin
(genome unpublished and not fully assembled)
The genome of a haploid Clementine orange (Citrus clementina) was sequenced by the International Citrus Genome Consortium to a coverage of 6.5-fold. The genome is not yet assembled into pseudomolecules but consists of 1,128 scaffolds containing a total of 296 megabases of sequence data. Genes were predicted using both sequencing of ESTs and homology to the genes of other sequenced plant species, resulting in a total of 25,385 protein coding genes. Download clementine sequence data and annotations from phytozome here (account creation/login required).
Papaya

The genome of the papaya tree (Carica papaya) was published in early 2008. Papaya was one of the earliest crops to be genetically modified (in papaya's case to resist the devastating papaya ringspot virus) and the sequenced genome actually comes from one of the genetically modified varieties (SunUp). The papaya genome was sequenced to a coverage of 3x using Sanger sequencing. Papaya has not experienced further whole genome duplications since the ancient hexaploidy shared by all currently sequenced eudicots. As the most closely related species to Arabidopsis with a currently sequenced genome that has not experienced the two subsequence whole genome duplications found in the Arabidopsis lineage, papaya is a useful outgroup, although the ancestors of Arabidopsis and Papaya split ~72 million years ago.
The papaya genome is estimated to have a size of 372 megabases, spread across nine chromosomes, and contain 28,629 genes. The version of papaya within CoGe is organized into super contigs, but does contain a number of gaps.
The genome paper:
Ming R et al. (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature, 452 (7190), 991-996 DOI: 10.1038/nature06856
Arabidopsis species and allies
Expect this category to grow substantially over the next year. The planned, in progress, and private genomes category below includes 7 more arabidopsis species and relatives.
Arabidopsis thaliana

Arabidopsis thaliana is a poplar model plant species, partially as a result of its short generation time and compact size. The genome of Arabidopsis was also the first plant genome to be published back in 2000. The current release of the Arabidopsis genome is TAIR10:
The TAIR10 release contains 27,416 protein coding genes, 4827 pseudogenes or transposable element genes and 1359 ncRNAs (33,602 genes in all, 41,671 gene models). A total of 126 new loci and 2099 new gene models were added.
The Arabidopsis genome is ~120 megabases of sequence spread across five chromosomes.
The Genome Paper: The Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408 (6814), 796-815 DOI: 10.1038/35048692
Genome resources:
- The 1001 genomes project[2] plans to sequence the genomes of 1001 different varieties of Arabidopsis. Currently 88 are available with more in progress. An analysis of the first 80 genomes was published in Nature Genetics in September 2011 (see the paper here).
Arabidopsis lyrata
Arabidopsis lyrata is a close relative of Arabidopsis thaliana. The ancestors of the two species split apart an estimated ten million years ago, making them somewhat closer than maize and sorghum among the grasses. A. lyrata is self-incompatable, while A. thaliana reproduced primarily through self-fertilization. The lyrata genome is also substantially larger than that of thaliana, weighing in at 207 MB, spread across seven chromosomes (compared to thaliana's 5 chromosomes and 125 megabase genome.)
The lyrata genome is available within CoGe, or you can download it from JGI.
Genome Paper: Tina T. Hu et al. (2011) "The Arabidopsis lyrata genome sequence and the basis of rapid genome size change." Nature Genetics 43:476–481 DOI: 10.1038/ng.807
Arabidopsis halleri
Unpublished but available under Ft. Lauderdale.
"Arabidopsis halleri is a perennial, outcrossing, insect-pollinated plant typically found 600m - 2300m above sea level in grassy meadows, forest margins and rocky crevices throughout Europe and eastern Asia. A. halleri has the unusual ability to colonize soil with high heavy metal content, which has made it a model for the study of heavy metal tolerance and accumulation in plants as well as understanding plant evolution and speciation in response to environmental change."
From phytozome [1] (where you can also download the current genome assembly after creating a JGI account).
Leavenworthia alabamica
"(i) Leavenworthia alabamica (lineage 1 in the tribe Camelineae), a model plant species with recently lost self-incompatibility in some populations;"
One of three genomes published simultaneously: An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions
Sisymbrium irio
"(ii) Sisymbrium irio (lineage 2 in the tribe Sisymbrieae), a self-compatible annual closely related to the Brassica genus but lacking the derived whole-genome triplication"
One of three genomes published simultaneously: An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions
Aethionema arabicum
"(iii) Aethionema arabicum (tribe Aethionemeae), a self-compatible, early branching sister group to the remainder of the core Brassicaceae"
One of three genomes published simultaneously: An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions
Brassica rapa
The genome of Brassica rapa was published in Nature Genetics in September 2011 by a consortum of researchers lead by the Beijing Genomics Institute (BGI). While the variety of Brassica rapa sequenced (Chiifu-401-42) is a breed of chinese cabbage, turnips are actually another cultivars of the same species. Brassica rapa is also one of the two parental species of Brassica napus an allotetraploid species which gives us both the vegetable rutabaga and the oil seed crop canola (also known as rapeseed, but seriously, who wants to buy a bottle named "Rape oil"?). Brassica rapa is the first corner of the Triangle of U to be sequenced. Since this publication, additional genome sequences have been published for Brassica oleracea and Brassica napus (and additional corner of the triable of U and the allotetraploid species formed by a cross between B. oleracea and B. rapa respectively.
In addition to the ancient hexaploidy shared by rosids and asterids and the two additional tetraploidies found in the shared Arabidopsis/Brassica lineage, the Brassica lineage experienced an additional ancient hexaploidy for a total duplication of 36 fold (3*2*2*3) relative to the pre-triplication common ancestor of the asterids and rosids.
Genome paper: The Brassica rapa Genome Sequencing Project Consortium. (2011) "The genome of the mesopolyploid crop species Brassica rapa." Nature Genetics DOI: 10.1038/ng.919
Brassica napus
Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome
"Oilseed rape (Brassica napus L.) was formed ~7500 years ago by hybridization between B. rapa and B. oleracea, followed by chromosome doubling, a process known as allopolyploidy. Together with more ancient polyploidizations, this conferred an aggregate 72× genome multiplication since the origin of angiosperms and high gene content. We examined the B. napus genome and the consequences of its recent duplication. The constituent An and Cn subgenomes are engaged in subtle structural, functional, and epigenetic cross-talk, with abundant homeologous exchanges. Incipient gene loss and expression divergence have begun. Selection in B. napus oilseed types has accelerated the loss of glucosinolate genes, while preserving expansion of oil biosynthesis genes. These processes provide insights into allopolyploid evolution and its relationship with crop domestication and improvement." --Science Abstract
Brassica oleracea
"Polyploidization has provided much genetic variation for plant adaptive evolution, but the mechanisms by which the molecular evolution of polyploid genomes establishes genetic architecture underlying species differentiation are unclear. Brassica is an ideal model to increase knowledge of polyploid evolution. Here we describe a draft genome sequence of Brassica oleracea, comparing it with that of its sister species B. rapa to reveal numerous chromosome rearrangements and asymmetrical gene loss in duplicated genomic blocks, asymmetrical amplification of transposable elements, differential gene co-retention for specific pathways and variation in gene expression, including alternative splicing, among a large number of paralogous and orthologous genes. Genes related to the production of anticancer phytochemicals and morphological variations illustrate consequences of genome duplication and gene divergence, imparting biochemical and morphological variation to B. oleracea. This study provides insights into Brassica genome evolution and will underpin research into the many important crops in this genus."
The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes
Raphanus raphanistrum
Wild radish. "Polyploidization events are frequent among flowering plants, and the duplicate genes produced via such events contribute significantly to plant evolution. We sequenced the genome of wild radish (Raphanus raphanistrum), a Brassicaceae species that experienced a whole-genome triplication event prior to diverging from Brassica rapa. Despite substantial gene gains in these two species compared with Arabidopsis thaliana and Arabidopsis lyrata, ∼70% of the orthologous groups experienced gene losses in R. raphanistrum and B. rapa, with most of the losses occurring prior to their divergence. The retained duplicates show substantial divergence in sequence and expression. Based on comparison of A. thaliana and R. raphanistrum ortholog floral expression levels, retained radish duplicates diverged primarily via maintenance of ancestral expression level in one copy and reduction of expression level in others. In addition, retained duplicates differed significantly from genes that reverted to singleton state in function, sequence composition, expression patterns, network connectivity, and rates of evolution. Using these properties, we established a statistical learning model for predicting whether a duplicate would be retained postpolyploidization. Overall, our study provides new insights into the processes of plant duplicate loss, retention, and functional divergence and highlights the need for further understanding factors controlling duplicate gene fate."
"Consequences of Whole-Genome Triplication as Revealed by Comparative Genomic Analyses of the Wild Radish Raphanus raphanistrum and Three Other Brassicaceae Species." http://www.ncbi.nlm.nih.gov/pubmed/24876251
Capsella rubella
Capsella is "the closest well-characterized genus" to arabidopsis and in fact the plants look rather similar to the eye of someone who doesn't study arabidopsis for a living. The best known Capsella species (and the only one with its own wikipedia page as I write this) is Capsella bursa-pastoris, which goes by the common name "shepard's purse." However C. bursa-pastoris is a tetraploid with all the challenges to genome assembly and genetic analysis that entails. Rather than tangle with the tetraploid genetics of bursa-pastoris, JGI instead aimed its sequencers at the closely related sister species Capsella rubella which has a much better behaved diploid genome.
The current assembly of Capsella rubella was generated from 22x sequencing with 454 reads, includes 134 megabases of assembled sequence, and has been assembled to the level of scaffolds. Genes were annotated using alignment of RNA-seq data from Capsella and homology to the genes of other sequenced eudicots.
The current assembly of the Capsella rubella genome is available from phytozome (account creation/login required).
Thellungiella parvula
Most people hadn't heard of this relative of arabidopsis prior to the publication of its genome in August 2011. Thellungiella, which goes by the common name "salt cress" is of interest because of its greatly increased tolerance for abiotic stresses (salt, cold, etc) relative to its much better studied relative Arabidopsis thaliana. The genome paper reported a genome approximately 140 megabases in size, assembled into seven pseudomolecules and emphasized the role of tandem duplicates in driving the remarkable stress tolerance of this species.
Thellungiella resources:
Genome paper: Maheshi Dassanayake et al (2011) "The genome of the extremophile crucifer Thellungiella parvula." Nature Genetics DOI: 10.1038/ng.889
Thellungiella/Eutrema Eutrema salsugineum
A second Thellungiella species was independently sequenced by the Chinese Academy of Sciences. The genome was sequenced to a depth of 134-fold coverage using Illumina sequencing.
Genome Paper: Hua-Jun Wu et al (2012) "Insights into salt tolerance from the genome of Thellungiella salsuginea." PNAS DOI: 10.1073/pnas.1209954109
A second version of the genome from the same species (sequenced using Sanger technology) was published by a group from JGI and the University of Arizona. This paper assigned the species to a different genus "Eutrema"
Genome Paper: Yang R et al (2013) "The reference genome of the halophytic plant Eutrema salsugineum" Frontiers in Plant Science DOI: 10.3389/fpls.2013.00046
For more details visit phytozome's page on the genome.
Hi Eric, David Jarvis in my laboratory was recently viewing the CoGePedia list of sequenced plant genomes, and noticed some issues with the Thellungiella species that I think we can help resolve. As it turns out, the Eutrema salsugineum (Yang et al 2013) genome is the same as the Thellungiella halophila (JGI) genome (all our work). When we began the genome project with JGI several years ago, everyone in the research community referred to the species as Thellungiella halophila. It was later noted that this species was not really Thellungiella halophila, but was in fact Thellungiella salsuginea. In addition, the whole genus was transferred to Eutrema, so the species is now known as Eutrema salsugineum. Thus, the Thellungiella halophila genome listed on Phytozome is really the Eutrema salsugineum genome sequence that was recently published by Yang et al. We have contacted JGI to see if they will change the name on Phytozome, and we thought we would also let you know in case you want to update the sequenced genomes list on CoGePedia. Will you please let me know if we can answer any questions or help in any way? Thanks and best regards. Karen Karen S. Schumaker, Ph.D. Interim Director and Professor, School of Plant Sciences Professor, Department of Molecular and Cellular Biology University of Arizona (April 21, 2013)
Cleome sp
The Cleome genus is an outgroup to all the other species contained within the "arabidopsis and allies" category. Two genome sequencing projects are currently in progress. Cleome hassleriana is being sequenced by a group involving Eric Schranz at Wageningen University and Cleome violacea is being sequenced by JGI. Despite sharing the same genus these species are actually well diverged (comparable to the evolutionary distance between Arabidopsis and Brassica) so comparisons of the two genome assemblies should be quite interesting.
Monocots
Duckweed (Spirodela polyrhiza)
"The subfamily of the Lemnoideae belongs to a different order than other monocotyledonous species that have been sequenced and comprises aquatic plants that grow rapidly on the water surface. Here we select Spirodela polyrhiza for whole-genome sequencing. We show that Spirodela has a genome with no signs of recent retrotranspositions but signatures of two ancient whole-genome duplications, possibly 95 million years ago (mya), older than those in Arabidopsis and rice. Its genome has only 19,623 predicted protein-coding genes, which is 28% less than the dicotyledonous Arabidopsis thaliana and 50% less than monocotyledonous rice. We propose that at least in part, the neotenous reduction of these aquatic plants is based on readjusted copy numbers of promoters and repressors of the juvenile-to-adult transition. The Spirodela genome, along with its unique biology and physiology, will stimulate new insights into environmental adaptation, ecology, evolution and plant development, and will be instrumental for future bioenergy applications." -- Abstract Nature Communications
Date Palm
In addition to being the first non-grass monocot genome to be published (in May of 2011 in the journal Nature Biotechnology, eight years after the first grass genome, rice), the paper describing the date palm (Phoenix dactylifera) genome is also the first scientific paper to pop up when you search of "Phoenix Genome." The current genome assembly includes 380 Megabases of sequence, which is only an estimated 60% of the total date palm genome, although it may include 90% of the gene space. Date palms have two sexes with separate make and female trees. Only the females produce dates so one of the key goals of the genome project was to be able to identify genetic tests to distinguish male and female seedlings, rather than having to wait 5-8 years for the plants to flower -- at which point it becomes obvious which plants are male and which are female.
Download link (at Weill Cornell Medical College).
Genome paper: Eman K Al-Dous et al., (2011) "De novo genome sequencing and comparative genomics of date palm (Phoenix dactylifera)." Nature Biotechnology 29:521–527 DOI: 10.1038/nbt.1860
Oil Palm
"Oil palm is the most productive oil-bearing crop. Although it is planted on only 5% of the total world vegetable oil acreage, palm oil accounts for 33% of vegetable oil and 45% of edible oil worldwide, but increased cultivation competes with dwindling rainforest reserves. We report the 1.8-gigabase (Gb) genome sequence of the African oil palm Elaeis guineensis, the predominant source of worldwide oil production. A total of 1.535 Gb of assembled sequence and transcriptome data from 30 tissue types were used to predict at least 34,802 genes, including oil biosynthesis genes and homologues of WRINKLED1 (WRI1), and other transcriptional regulators1, which are highly expressed in the kernel. We also report the draft sequence of the South American oil palm Elaeis oleifera, which has the same number of chromosomes (2n = 32) and produces fertile interspecific hybrids with E. guineensis2 but seems to have diverged in the New World. Segmental duplications of chromosome arms define the palaeotetraploid origin of palm trees. The oil palm sequence enables the discovery of genes for important traits as well as somaclonal epigenetic alterations that restrict the use of clones in commercial plantings3, and should therefore help to achieve sustainability for biofuels and edible oils, reducing the rainforest footprint of this tropical plantation crop."
Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds
Banana
The banana genome was published on June 11th 2012. The banana genome is currented based on a doubled haploid individual of Musa acuminata not the triploid Cavendish bananas which are a familiar sight in in grocery stores around the world. The genome was sequenced to a depth of 20x using a combination of 454 and Sanger sequencing, and 50x using Illumina short read sequencing technology. The 11 pseudomolecules of the current assembly cover 332 megabases (63% of the estimated total banana genome size of 523 megabases). An additional 140 megabases of unanchored scaffolds and contigs (33% of the estimated total genome size) are also included in the current genome release. While delays prevented banana from being the first non-grass monocot genome to be published (Date Palm claimed that title), the authors of the banana paper do say theirs is the "...first monocotyledon high-continuity whole-genome sequence reported outside Poales...."
The argument for sequencing banana isn't to make the lives of comparative genomicists easier, but because of the key role many banana species play in the tropical food production. Bananas are also a target of genetic engineering, since the fact that most cultivated breeds are triploid and unable to reproduce sexually (the reason bananas aren't full of seeds) makes conventional breeding extremely difficult (although that doesn't stop some dedicated biologists from doing it anyway), and bananas suffer from a number of nasty plant pathogens.
Trivia: The banana is the most consumed fruit in the United States, with the average American eating ~25 pounds of bananas per year, a full quarter of the average total fruit consumed per person. Yet almost NO bananas are produced domestically in the USA.
The Genome Paper: Angelique D'Hont et al (2012) "The banana (Musa acuminata) genome and the evolution of monocotyledonous plants." Nature DOI: 10.1038/nature11241
Grasses
The grasses, a family of plants known as the poaceae, can trace their lineages back to a common ancestor that probably lived between 50-70 million years ago, either right before or soon after the extinction of the dinosaurs(dinosaurs didn't eat grass). Since their emergence in the fossil record, the grasses have been extraordinarily successful, becoming one of the largest families of plants on the planet and covering vast swaths of the planet in the form of prairies/savannahs/steppes.
While you may think of grass primarily as the green stuff on lawns and sports fields, remember that grasses also include species like bamboo and the grains that make up so much of what we eat. Either three (rice, wheat, and corn/maize) or four (the same three plus sugar cane) grass species provide more than half of all the calories that feed the worlds population[3], and are the focus of much applied and basic scientific research. Check out the Pan-grass synteny project
Rice and rice relatives
Asian rice

Rice (Oryza sativa) was the second plant genome (after Arabidopsis) to be published, making it the first monocot genome, the first grass genome, the first food crop genome, and the first grain genome (and probably a whole lot of other firsts as well). The original published genome (published in 2002) was actually a dual publication by two independent groups, one of the subspeciesjaponica and another of the subspeciesindica.
The current version of the rice genome in CoGe is v6.1 from MSU (the japonica version of the genome) which contains ~370 megabases of sequence and 40,577 non-transposon related genes spread across 12 chromosomes.
Rice Resources:
- The MSU Rice Gene Annotation Project the source of the gene annotations used by CoGe
- RAP-DB: The Rice Annotation Project Database. A different set of annotations for the rice genome
The genome paper:
Goff, S. et al. (2002). A Draft Sequence of the Rice Genome (Oryza sativa L. ssp. japonica) Science, 296 (5565), 92-100 DOI: 10.1126/science.1068275
Yu, J. et al. (2002) A Draft Sequence of the Rice Genome (Oryza sativa L. ssp. indica) Science 2006 (5565), 79-92 DOI: 10.1126/science.1068037
The wild progenitor of Asian rice is Oryza rufipogon which has [its own genome project! http://plants.ensembl.org/Oryza_rufipogon/Info/Index]. Note that this is an unpublished Ft. Lauderdale restrained dataset.
African Rice
The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication
"The cultivation of rice in Africa dates back more than 3,000 years. Interestingly, African rice is not of the same origin as Asian rice (Oryza sativa L.) but rather is an entirely different species (i.e., Oryza glaberrima Steud.). Here we present a high-quality assembly and annotation of the O. glaberrima genome and detailed analyses of its evolutionary history of domestication and selection. Population genomics analyses of 20 O. glaberrima and 94 Oryza barthii accessions support the hypothesis that O. glaberrima was domesticated in a single region along the Niger river as opposed to noncentric domestication events across Africa. We detected evidence for artificial selection at a genome-wide scale, as well as with a set of O. glaberrima genes orthologous to O. sativa genes that are known to be associated with domestication, thus indicating convergent yet independent selection of a common set of genes during two geographically and culturally distinct domestication processes." --Nature Genetics Abstract
Wild ancestor is Oryza barthii which also has a [prepublication genome dataset http://plants.ensembl.org/Oryza_barthii/Info/Index].
Assorted other Oryza
In additiom to wild and domesticated species for African and Asian rice, there are also [pre-publication datasets for five more species in the genus Oryza http://plants.ensembl.org/Multi/Search/Results/species_plants?db=core;q=oryza;species=all;collection=all;site=ensemblunit]:
- Oryza nivara
- Oryza glumaepatula
- Oryza punctata
- Oryza brachyantha
- Oryza meridionalis
[OMAP project http://www.omap.org/] has so far
Brachypodium

The brachy genome (Brachypodium distachyon) was published in early 2010. Brachypodium is a small temperate grass native found around the Mediterranean, and east into India. Its choice as a model organism was based on small physical size, quick generation time, and small genome (a lot of the same reasons as Arabidopsis) as well as its membership in the Pooideae, a group of grass species that also includes important crop species: wheat, barley, rye, and oats all species whose genomes have not yet been sequenced (although the last common ancestor of brachy and these important crop species is estimated to have lived >30 million years ago). Brachy's genome is currently the only published genome of a non-domesticated grass and the only temperate (as opposed to tropical) grass.
The published version of the brachy genome includes 272 megabases of sequence and 25,532 protein coding genes spread across five chromosomes. It was sequenced to a coverage of 9.4× using Sanger sequencing.
Brachy Resources:
- Brachypodium.org is probably the first place to check.
- Brachy's page at JGI (account creation/login required)
The genome paper:
Vogel, J et al (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon Nature, 463 (7282), 763-768 DOI: 10.1038/nature08747
Barley
Barley (Hordeum vulgare) is an important crop species around the world, a relative of wheat, and currently the single largest plant genome sequenced to date (weighing in at a haploid genome size of 5.1 gigabases, more than twice the size of the next biggest sequenced genome maize). Barley is the fourth most widely grown grain in the world behind the big three: rice, maize, and wheat. In addition to being used in soups, breads, and as animal feed, barley is perhaps most widely known as at source of the malt used to produce most beer around the world.
The barley genome was assembled using a mixture of BAC-sequencing, anchoring to a high density genetic map, and syntenic path assembly.
The genome paper:
The International Barley Genome Sequencing Consortium (2012) A physical, genetic and functional sequence assembly of the barley genome Nature doi: 10.1038/nature11543
Wheat and wheat-like substances
Einkorn wheat
Einkorn wheat (Triticum urartu) is closely related to the "A" genome found in hexaploid bread wheats (ABD genome) and tetraploid durum wheat (AB). Einkorn wheat has a genome size of ~5 gigabases and the genome was sequenced using short read Illumina technology. While a majority of the genome is likely represented in the assembly (3.92 gigs of assembled sequence), it should be noted that half the genome is found in scaffolds of less than 64 kilobases. <-- which is still perfectly respectable for sequencing an 5 gigabase genome using short reads.
The Genome Paper Ling HQ et al (2013) "Draft genome of the wheat A-genome progenitor Triticum urartu" Nature DOI: 10.1038/nature11997
Moso Bamboo
"Bamboo" refers to a whole clade of giant woody grass species. Despite the economic and ecological significance of these species the large genome sizes of bamboos and the difficulty of carrying out genetics (different bamboo species flower only every 65-130 years) made obtaining a bamboo reference genome difficult. However in 2013, a Chinese research group published a draft genome of moso bamboo (Phyllostachys heterocycla), a species which apparently accounts for 70% of bamboo forests.
The genome was assembled using a primarily next generation sequencing approach, and covers an estimated 88% of the bamboo genome (70+% of the genome in scaffolds longer than 65 KB). The researchers identified a whole genome duplication which occurred in bamboo between 7-12 million years ago.
Download the bamboo genome and annotations here: http://www.ncgr.ac.cn/bamboo
The Genome Paper: Peng Z et al (2013) "The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla)." Nature Genetics doi: [10.1038/ng.2569
Maize/Corn

The genome of the species known to most Americans as corn (Zea mays) and to biologists and Europeans as maize was published in the second half of 2009. Maize genetics has a history going back more than a century to the early work R. A. Emerson, widely considered the founder of modern maize genetics. Maize is an important crop species, and the most prominent crop species to engage in C4 photosynthesis (as opposed to the more standard C3 photosynthesis). The role of maize as an important model system as well as a vital crop might have placed it earlier in the order of plants to have their genomes sequenced if not for the complexity of the genome itself.
The ancestor of maize went through a Whole genome duplication between 5 and 12 million years ago. In additio, the recent history of maize has included not one but two blooms of transposon activity. The result is a genome that weighs in at ~2.5 gigabases of mostly repetitive sequence, making both sequencing and assembly major challenges.
But the maize genome sequence is now published.
The v1 sequence contains 2.3 gigabases of sequence data. Rather than shotgun sequencing of the entire genome as is now common with smaller less repetitive genomes, maize was sequenced using a BAC[4] by BAC approach. The BACs were lined up to cover the ten chromosomes of maize, and then the sequence contained in each BAC was shotgun sequenced and assembled into contigs. What this means in practice is that a given sequence in the maize genome is usually within 300 kilobases of its correct location, but within that range may be out of order or inverted. If a gene seems to be absent from its syntenic location (or only a portion of the gene is found) it is important to search up to 500 kilobases in either direction around its expected location to make sure the apparent deletion isn't the result of incorrect ordering of the contigs.
This issue was reduced in version 2 of the genome released in the spring of 2010 as over 80% of the contigs in the v2 sequence have data on their order and orientation, up from ~30% in the v1 release.
A word on gene models:
The maize genome was published with two sets of genome annotations, the working gene set and the filtered gene set. These two sets are based on different compromises between catching all the real genes in maize and excluding false genes.
- The filtered gene set (>32,000 genes (this number is from version 1, there are more in the version 2 filtered gene set released in February 2011)) are high confidence genes. If it's in the filtered gene set, it's almost certainly a gene, but there is no promise that EVERY real gene is in the filtered gene get
- The working gene set (~100,000 genes) includes all the genes in the filtered gene set, but also many other gene models that have less supporting gene evidence. Almost every real gene is likely included in the working gene set, but so are many things that aren't genes, particularly gene fragments remaining from the maize whole genome duplication, and pieces of genes captured by transposons.
The maize genome is divided among 10 chromosomes.
Maize Resources:
Maize Related CoGepedia Pages:
- Classical Maize Genes: ~460 maize genes that we have manually mapped to gene models in the published genome sequence, plus data on syntenic orthologs in rice, sorghum, and brachy, as well as the homeologous region of maize.
- MaizeGDB and CoGe: Explaining how to jump between our site and MaizeGDB
- Maize Sorghum Syntenic dotplot: How to compare the maize and sorghum genomes.
The genome paper:
Schnable, P et al (2009) The B73 Maize Genome: Complexity, Diversity, and Dynamics Science, 326(5956), 1112-1115 DOI: 10.1126/science.1178534
Companion issue in PLoS Genetics published simultaneously with the genome paper:
PLoS Genetics: 2009 Maize Genome Collection
Sorghum

Sorghum (Sorghum bicolor) is an important grain species. A close relative of maize, sorghum is generally considered to be an even more stress tolerant crop. Like maize it carries out C4 photosynthesis. It does not share the recent whole genome duplication seen in maize, which makes it an excellent outgroup from studies of that event in maize as the common ancestor of maize and sorghum is estimated to have lived only 12 million years ago.
The sorghum genome was published in 2009. The current version in CoGe (v1.4) contains ~700 megabases of sequence and 34,496 protein coding genes spread over ten chromosomes. The sorghum genome sequence is available from phytozome (account creation/login required).
The genome paper: Paterson, A et al. (2009) The Sorghum bicolor genome and the diversification of grasses Nature, 457 (7229), 551-556 DOI: 10.1038/nature07723
Foxtail Millet
Foxtail Millet (Setaria italica) is a C4 grass. It is the first species in Paniceae, a tribe of grasses that includes switchgrass and is sister to the Andropogoneae (the tribe that maize and sorghum belong to), to have its genome sequenced. The Paniceae also contain switchgrass, an important biofuel crop with an intractable genome, which was one of the several justifications for the sequencing the foxtail millet genome. Foxtail millet was domesticated in China and is a more distant relative to maize and sorghum.
The current version of the Setaria genome loaded into CoGe (v2.1) includes 406 megabases of sequence and 35,471 annotated genes.
Two versions of the foxtail millet genome were generated independently by the Beijing Genomics Institute and the Joint Genomes Institute and published in the same issue of Nature Biotechnology:
Zheng G et al. (2012) "Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential." Nature Biotechnology doi: 10.1038/nbt.2195
Bennetzen J et al. (2012) "Reference genome sequence of the model plant Setaria." Nature Biotechnology doi: 10.1038/nbt.2196
Tef (Eragrostis tef)
"Tef (Eragrostis tef), an indigenous cereal critical to food security in the Horn of Africa, is rich in minerals and protein, resistant to many biotic and abiotic stresses and safe for diabetics as well as sufferers of immune reactions to wheat gluten. We present the genome of tef, the first species in the grass subfamily Chloridoideae and the first allotetraploid assembled de novo. We sequenced the tef genome for marker-assisted breeding, to shed light on the molecular mechanisms conferring tef’s desirable nutritional and agronomic properties, and to make its genome publicly available as a community resource." --BMC Genomics Abstract
Non-angiosperms
Gymnosperms
Norway Spruce
"Conifers have dominated forests for more than 200 million years and are of huge ecological and economic importance. Here we present the draft assembly of the 20-gigabase genome of Norway spruce (Picea abies), the first available for any gymnosperm. The number of well-supported genes (28,354) is similar to the >100 times smaller genome of Arabidopsis thaliana, and there is no evidence of a recent whole-genome duplication in the gymnosperm lineage. Instead, the large genome size seems to result from the slow and steady accumulation of a diverse set of long-terminal repeat transposable elements, possibly owing to the lack of an efficient elimination mechanism. Comparative sequencing of Pinus sylvestris, Abies sibirica, Juniperus communis, Taxus baccata and Gnetum gnemon reveals that the transposable element diversity is shared among extant conifers. Expression of 24-nucleotide small RNAs, previously implicated in transposable element silencing, is tissue-specific and much lower than in other plants. We further identify numerous long (>10,000 base pairs) introns, gene-like fragments, uncharacterized long non-coding RNAs and short RNAs. This opens up new genomic avenues for conifer forestry and breeding."
The Norway spruce genome sequence and conifer genome evolution In Nature in 2013
Loblolly Pine
"Background The size and complexity of conifer genomes has, until now, prevented full genome sequencing and assembly. The large research community and economic importance of loblolly pine, Pinus taeda L., made it an early candidate for reference sequence determination. Results We develop a novel strategy to sequence the genome of loblolly pine that combines unique aspects of pine reproductive biology and genome assembly methodology. We use a whole genome shotgun approach relying primarily on next generation sequence generated from a single haploid seed megagametophyte from a loblolly pine tree, 20-1010, that has been used in industrial forest tree breeding. The resulting sequence and assembly was used to generate a draft genome spanning 23.2 Gbp and containing 20.1 Gbp with an N50 scaffold size of 66.9 kbp, making it a significant improvement over available conifer genomes. The long scaffold lengths allow the annotation of 50,172 gene models with intron lengths averaging over 2.7 kbp and sometimes exceeding 100 kbp in length. Analysis of orthologous gene sets identifies gene families that may be unique to conifers. We further characterize and expand the existing repeat library based on the de novo analysis of the repetitive content, estimated to encompass 82% of the genome. Conclusions In addition to its value as a resource for researchers and breeders, the loblolly pine genome sequence and assembly reported here demonstrates a novel approach to sequencing the large and complex genomes of this important group of plants that can now be widely applied."
Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies Genome Biology in 2014
Physcomitrella patens
Physcomitrella patens is a moss. Mosses, along with liverworts and hornworts, make up the bryophytes a group of plants that have neither flowers nor vascular tissue. We think bryophytes still look a lot like the ancestors of all land plants, but it is important to remember that bryophytes alive today, like Physcomitrella patens, have been evolving from that common ancestor for just as long as rice or arabidopsis. ~450 million years in all three cases.
The Physcomitrella genome was published in early 2008 and consists of 480 megabases of sequence and 35,938 gene models spread over 2,106 scaffolds. (Physcomitrella has 27 chromosomes.) The genome was sequenced to a depth of 8x coverage using Sanger shotgun sequencing.
Physcomitrella resources:
- The genome is available from JGI.
- More links from mossgenome.org
The genome paper:
Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, Tanahashi T, Sakakibara K, Fujita T, Oishi K, Shin-I T, Kuroki Y, Toyoda A, Suzuki Y, Hashimoto S, Yamaguchi K, Sugano S, Kohara Y, Fujiyama A, Anterola A, Aoki S, Ashton N, Barbazuk WB, Barker E, Bennetzen JL, Blankenship R, Cho SH, Dutcher SK, Estelle M, Fawcett JA, Gundlach H, Hanada K, Heyl A, Hicks KA, Hughes J, Lohr M, Mayer K, Melkozernov A, Murata T, Nelson DR, Pils B, Prigge M, Reiss B, Renner T, Rombauts S, Rushton PJ, Sanderfoot A, Schween G, Shiu SH, Stueber K, Theodoulou FL, Tu H, Van de Peer Y, Verrier PJ, Waters E, Wood A, Yang L, Cove D, Cuming AC, Hasebe M, Lucas S, Mishler BD, Reski R, Grigoriev IV, Quatrano RS, Boore JL. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319 (5859):64-9 DOI: 10.1126/science.1150646
Selaginella moellendorffii
Selaginella moellendorffii is a lycophyte, an ancient branch of the plant tree of life. Like mosses, lycophytes do not have flowers, but lycophytes do have a vascular system. Lycophytes are often grouped with ferns as vascular non-seed producing plants. Selaginella has the distinction of currently being the smallest sequenced plant genome (~110 megabases, smaller than Arabidopsis!) and having a dedicated wiki.
Less genomics related, but exciting never the less is that plants that recognizably belong to the Selaginella genus can be found for the last 335-350 million years in the fossil record. That is older than the dinosaurs!
Selaginella resources:
- Selaginella wiki
- Jo Ann Banks wrote a great review on Selaginella which can be found here.
- JGI page where you can download the v1.0 assembly of the Selaginella genome.
Genome paper: Jo Ann Banks et al. (2011) "The Selaginella Genome Identifies Genetic Changes Associated with the Evolution of Vascular Plants." Science 332:960-963 DOI: 10.1126/science.1203810
Green Algae
Chlamydomonas reinhardtii
Single celled chlorophyte (green alga) found all over the world in many different environments. Important model organism due to its photosynthetic capabilities, methods for genetically modifying it, and short generation time. In addition, these traits make Chlamydomonas a strong candidate as a source for biofuels.
Genome published in Science: http://www.sciencemag.org/content/318/5848/245.short
Available from the Joint Genome Institute: http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Creinhardtii (account creation/login required)
Volvox carteri
"The multicellular green alga Volvox carteri and its morphologically diverse close relatives (the volvocine algae) are well suited for the investigation of the evolution of multicellularity and development. We sequenced the 138-mega-base pair genome of V. carteri and compared its approximately 14,500 predicted proteins to those of its unicellular relative Chlamydomonas reinhardtii. Despite fundamental differences in organismal complexity and life history, the two species have similar protein-coding potentials and few species-specific protein-coding gene predictions. Volvox is enriched in volvocine-algal-specific proteins, including those associated with an expanded and highly compartmentalized extracellular matrix. Our analysis shows that increases in organismal complexity can be associated with modifications of lineage-specific proteins rather than large-scale invention of protein-coding capacity."
Published in Science: http://www.ncbi.nlm.nih.gov/pubmed/20616280
http://www.sciencemag.org/lookup/pmid?view=long&pmid=20616280
Available from JGI: http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Vcarteri
Ostreococcus lucimarinus
"The smallest known eukaryotes, at ≈1-μm diameter, are Ostreococcus tauri and related species of marine phytoplankton. The genome of Ostreococcus lucimarinus has been completed and compared with that of O. tauri. This comparison reveals surprising differences across orthologous chromosomes in the two species from highly syntenic chromosomes in most cases to chromosomes with almost no similarity. Species divergence in these phytoplankton is occurring through multiple mechanisms acting differently on different chromosomes and likely including acquisition of new genes through horizontal gene transfer. We speculate that this latter process may be involved in altering the cell-surface characteristics of each species. In addition, the genome of O. lucimarinus provides insights into the unique metal metabolism of these organisms, which are predicted to have a large number of selenocysteine-containing proteins. Selenoenzymes are more catalytically active than similar enzymes lacking selenium, and thus the cell may require less of that protein. As reported here, selenoenzymes, novel fusion proteins, and loss of some major protein families including ones associated with chromatin are likely important adaptations for achieving a small cell size."
[The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation http://www.pnas.org/content/104/18/7705.long] Published in PNAS in 2007
Genome is available from Phytozome: http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Olucimarinus
Bathycoccus prasinos
"Background Bathycoccus prasinos is an extremely small cosmopolitan marine green alga whose cells are covered with intricate spider's web patterned scales that develop within the Golgi cisternae before their transport to the cell surface. The objective of this work is to sequence and analyze its genome, and to present a comparative analysis with other known genomes of the green lineage.
Research Its small genome of 15 Mb consists of 19 chromosomes and lacks transposons. Although 70% of all B. prasinos genes share similarities with other Viridiplantae genes, up to 428 genes were probably acquired by horizontal gene transfer, mainly from other eukaryotes. Two chromosomes, one big and one small, are atypical, an unusual synapomorphic feature within the Mamiellales. Genes on these atypical outlier chromosomes show lower GC content and a significant fraction of putative horizontal gene transfer genes. Whereas the small outlier chromosome lacks colinearity with other Mamiellales and contains many unknown genes without homologs in other species, the big outlier shows a higher intron content, increased expression levels and a unique clustering pattern of housekeeping functionalities. Four gene families are highly expanded in B. prasinos, including sialyltransferases, sialidases, ankyrin repeats and zinc ion-binding genes, and we hypothesize that these genes are associated with the process of scale biogenesis.
Conclusion The minimal genomes of the Mamiellophyceae provide a baseline for evolutionary and functional analyses of metabolic processes in green plants."
Gene functionalities and genome structure in Bathycoccus prasinos reflect cellular specializations at the base of the green lineage Genome Biology in 2012
Klebsormidium flaccidum
"The colonization of land by plants was a key event in the evolution of life. Here we report the draft genome sequence of the filamentous terrestrial alga Klebsormidium flaccidum (Division Charophyta, Order Klebsormidiales) to elucidate the early transition step from aquatic algae to land plants. Comparison of the genome sequence with that of other algae and land plants demonstrate that K. flaccidum acquired many genes specific to land plants. We demonstrate that K. flaccidum indeed produces several plant hormones and homologues of some of the signalling intermediates required for hormone actions in higher plants. The K. flaccidum genome also encodes a primitive system to protect against the harmful effects of high-intensity light. The presence of these plant-related systems in K. flaccidum suggests that, during evolution, this alga acquired the fundamental machinery required for adaptation to terrestrial environments."
Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation Nature Communications 2014
Planned, In-progress, and Private genome sequencing efforts (a partial list)
- The genome of the dwarf birch tree Betula nana is currently being assembled by Richard Buggs at the university of London.
- The sunflower genome project was just announced in early 2010. While it's far too early to predict when this genome will be released, it is still worth mentioning, because species within the sunflower genus (Helianthus) have genome sizes around 3000 megabases (sometimes substantially more) making this genome a candidate to steal from maize/corn the position of largest sequenced plant genome. More information here (warning this is a pdf formatted press release)
- Bayer CropScience announced they have a complete genome sequence for canola (Bassica napus) as well as varieties of Brassica rapa and Brassica oleracea. These aren't being released publicly, but from what I've heard they are open to collaborating with individual researchers who want access to the data.
- Several different companies have announced that they have sequenced the genome of the oil palm, but to the best of our knowledge none of these sequences are publicly available. News reports of the sequencing:
- Seed plant genomes listed by JGI (the Joint Genome Institute, part of the US Department of Energy) in approximate order of progress (as best I can tell I've listed those closest to completion at the top):
- Boechera stricta Arabidopsis relative
- Seagrass (Zostera marina) a monocot.
- Loblolly Pine (Pinus taeda)
- Arabidopsis halleri Arabidopsis species
- Boechera holboellii Arabidopsis relative
- Miscanthus giganteus a biofuel crop not unlike switchgrass
- Panic grass (Panicum hallii) a switchgrass relative
- Arabidopsis arenosa an arabidopsis species
- Boechera divericarpa an arabidopsis relative
- Switchgrass (Panicum virgatum)
- Purple willow (Salix purpure)
- Fruit tree genomes currently in the process of being sequenced/assembled/annotated by researchers including Amit Dhingra's group at Washington State:
- Pear (Pyrus communis (?) )
- Sweet Cherry (Prunus avium)
- A group of researchers are currently in the process of sequencing the genome of Common Milkweed (Asclepias syriaca) using Illumina short read sequencing.
- Other Genomes in progress culled from abstracts from the 2011 Plant and Animal Genome Conference
- Barley
- African Cultivated Rive (Oryza glaberrima)
- Coffee (Coffea caneophora)
- Lettuce
- Watermelon
- Chinese Chestnut (Castanea mollissima)
- European poplar (Populus nigra)
- European aspen (Populus tremula)
- European plum (Prunus domestica)
- Red Raspberry (Rubus idaeus cv. Heritage)
- Sangiovese Grapevine (Vitis vinifera var Sangiovese)
- Peanut (Arachis hypogaea)
Citations to this page
Have you used the information gathered together on this page to assist you in writing a publication? Please let us know! It's a lot of work trying to keep all these entries up to date and the rewarding part is hearing how the final result is put to good use.
Here are some publications which have used the sequenced plant genomes page (publications of the Freeling & Lyons lab omitted):
- Shi J et al (2013) "Evolutionary Dynamics of Microsatellite Distribution in Plants: Insight from the Comparison of Sequenced Brassica, Arabidopsis and Other Angiosperm Species" doi:10.1371/journal.pone.0059988
- Nelson DR & Schuler MA (2013) "Cytochrome P450 Genes from the Sacred Lotus Genome" http://link.springer.com/article/10.1007/s12042-013-9119-z
- Kelly LJ (2012) "Why size really matters when sequencing plant genomes" doi: 10.1080/17550874.2012.716868
- Ranjan A et al (2012) "The tomato genome: implication for plant breeding, genomics, and evolution." Genome Biology doi:10.1196/gb-2012-13-8-167
- Bowers JE et al (2012) "Development of a 10,000 Locus Genetic Map of the Sunflower Genome Based on Multiple Crosses." G3 doi: 10.1534/g3.112.002659
- Salse J & Feuillet C (2011) "Palaeogenomics in cereals: Modeling of ancestors for modern species improvement." Comptes Rendus Biologies doi: 10.1016/j.crvi.2010.12.014
- Utomo HS et al (2012) "Progression of DNA Marker and the Next Generation of Crop Development." Crop Plant, Dr Aakash Goyal (Ed.), ISBN: 978-953-51-0527-5, InTech.
Footnotes
- ↑ 1.0 1.1 Groupings of dna sequence that correspond to the individual chromosomes of an organisms
- ↑ Literally one uping the 1000 genome project that plans to sequence the genomes of 1000 people
- ↑ Estimated to be 6.7 billion people as of early 2010
- ↑ Bacterial artificial chromosome. A way of break down a genome into managable chunks of ~300 kilobases