SynMap

Overview
SynMap allows you to generate a syntenic dotplot between two organisms and identify syntenic regions. This is done by:
- Finding putative genes or regions of homology between two genomes
- Identifying collinear sets of genes or regions of sequence similarity to infer synteny
- Generating a dotplot of the results and coloring syntenic pairs.
If you choose, synonymous and non-synonymous site mutation data can be calculated for protein coding genes that are identified as syntenic. These genes will then be colored based on those values in the dotplot for rapid identification of different age-classes of syntenic regions.
SynMap Methods
- Extract sequences for comparison; build fasta files
- Create blastable databases (if necessary) and compare using
- Identifies tandem gene duplicates using a program written by Haibao Tang and Brent Pedersen called blast2raw
- Filters repetitive matches using a program written for SynMap by Brent Pedersen
- Identify syntenic pairs of by finding collinear series of putative homologous sequences using DAGChainer [3]
- Optional: calculate synonymous and nonsynonymous mutation rates for syntenic gene pairs using CodeML of the PAML package[4]
- Generate dotplot of all putative homologous matches; dots are colored gray
- Add syntenic pairs; dots are colored either green and red (same or opposite orientation), or based on the synonymous mutations, nonsynonymous mutations, or the ratio of nonsynonymous/synonymous mutations
- ↑ Schwartz, S. et al. Human-mouse alignments with BLASTZ. Genome Res. 13, 103−107 (2003)
- ↑ Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. Journal of Molecular Biology 215:403-410
- ↑ Haas BJ, Delcher AL, Wortman JR, Salzberg SL (2004) DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics 20: 3643–3646
- ↑ Yang Z (2007) PAML 4: Phylogenetic Analysis by Maximum Likelihood. Molecular Biology and Evolution 24:1586-1591
Specifying genomes
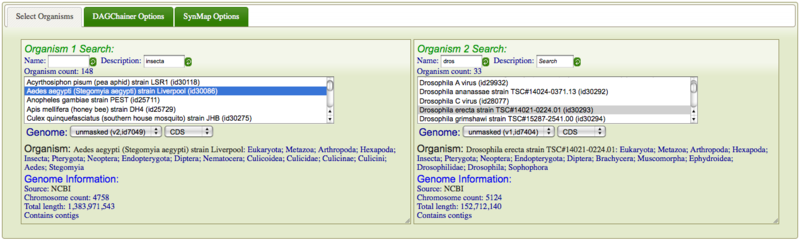
It is easy to search and select organisms in SynMap. Just select the "select organisms" tab and search for two organisms by either name or organism description. Just type part of either in the text box and SynMap automatically searches for any organism that contains your text. The results are displayed below the search boxes. Some organisms may have multiple versions of their genome and different types of sequences (e.g. masked versus unmasked). These will be displayed in a drop-down menu from which you can select the correct genome. Also, this is where you can select for comparing CDS sequence or genomic sequence. If the genome does not have CDS features, the option won't be display and a warning will be printed in below these drop-down menus. When selected, a brief description of the genome will be displayed below the drop-down menus. This will include the full organism name and description, followed by an overview of the genome. If you click on "Genome Information:" it will link to OrganismView and give you a full description of the genome. Otherwise, it will display source of the genome, the number of chromosomes in the genome, the total length of the genome in nucleotides, and whether the genome contains plasmids or contigs.
DAGChainer options
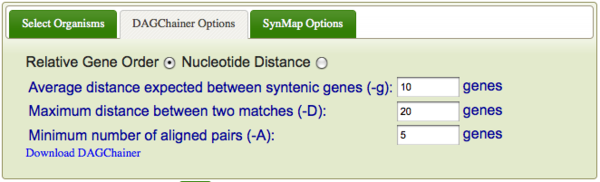
DAGChainer is the algorithm used for finding series of collinear genes (or regions of sequence similarity) between two genomes in order to infer synteny. To specify DAGChainer options, select the "DAGChainer Options" tab. DAGChainer works by searching some distance from a pair of genes for another pair. If some threshold number of gene pairs are identified, DAGChainer keeps that set of gene-pairs which will be reported back to SynMap. These sets of gene pairs are interpreted as two syntenic regions, and get colored in the dotplot for easy identification.
The options you can specify are:
- Are genomic coordinates in genes or nucleotide units ("Relative Gene Order" and "Nucleotide Distance" respectively).
- This determines whether DAGChainer is searching by number of genes (or regions of sequence similarity) or by absolute genomic nucleotide distances. For the most part, using "relative gene order" is preferable. The absolute distance between genes in nucleotides varies widely between genomes, and even within a genome. For the former, gene spacing in a bacteria genome are orders of magnitude closer than for an animal genome. For the latter, genes near centromeres are often further from one another than genes distal to a centromere. This is very apparent in plant genomes.
- Average distance expected between syntenic genes
- Maximum distance between two matches
- These two options are self explanatory. One thing to keep in mind is that the larger these values are set, the more generous DAGChainer is in including genes in a collinear set. In other words, as these values are increased, your false positive rate goes up. Also, the "average distance" should be lower than "maximum distance".
- Minimum number of aligned pairs
- This is the minimum number of gene-pairs that DAGChainer needs in a collinear gene set to keep. The higher this number, the more stringent DAGChainer will be for finding "good" collinear gene sets.
QuotaAlign options
QuotaAlign is a post-processing step to merge adjacent syntenic blocks, or select subset of syntenic blocks that reflect matching ratio of regions (for example, number of subgenomes in duplicated genomes). These two steps are essential for downstream analysis of genome rearrangements. Read more details on QuotaAlign.
SynMap options
- Blast Algorithm: blastn: nucleotide-nucleotide search; tblastx: translated nucleotide-translated nucleotide search
- tblastx takes 6 times longer than blastn and usually doesn't improve the ability to find synteny. If the DNA sequence is that diverged at the nucleotide level that a protein sequence search is needed to find putative homologous sequence, usually the genome structure is also very divergent and collinear gene sets are not likely to be found
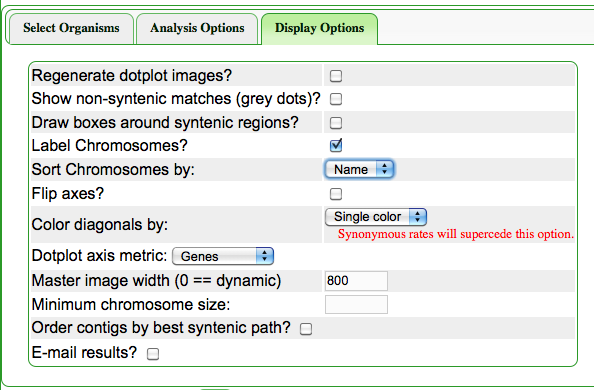
- Regenerate dotplot images: Select this if you want the dotplot image regenerated. This are saved and can be out of date with regards to the methods used to generate them, or something bad happened during their original generation
- Dotplot axus metric: Are the dotplot axis to be measured in nucleotide or gene units. Nucleotides accurately reflect the structure of the genome. Gene units help make collinear lines straighter if there are regions of the genome undergoing different rates of expansions (e.g. centromeres).
- Master image width: How wide, in pixels, will the master dotplot image be? Dynamic scales it according to the size of the genome, but sometimes it is necessary to make them very large to see some details.
- Calculate substitute rates for syntenic protein coding gene pairs and color syntenic dots accordingly
- This is the option to set if you want to calculate synonymous, nonsynonymous, or the ratio of those and color syntenic gene pair dots based on those values. This is very helpful if there are whole genome duplication events in one or both genomes. When these values are generated, a histogram of the values will also be generated and displayed. The histogram will be color coded identically to the dots in the dotplot. This makes it easy to determine which sytnenic gene pair dots are older or newer than other syntenic dots. For an example, please see below. PLEASE NOTE: generating these values takes a long time. It takes a very long time for large genomes.
- Order contigs by best syntenic path: this option will invoke a special algorithm to rearrange contigs in order to create the best order of contigs that make a continous syntenic path in relation to a reference genome. This is very useful for a genome sequences that have been assembled to contigs for which a reference genome is available. PLEASE NOTE: that this ordering of contigs may not be how the genome is actually structured. Some genomic changes, such as large-scale inversions will not be correctly placed if the contigs' break-points are at the end of the inversion. This is often the case as inversions happen in repetitive sequence, and these same repetitive sequence are often where genome assembly algorithms cannot assemble through. For an example, see Syntenic path assembly.
Calculating and displaying synonymous/non-synonymous (Ks, Kn) data
This option is selected under the "SynMap Options" tab To select, just set the option "Calculate substitution rates for syntenic protein coding gene pairs and color syntenic dots accordingly" to what rate you desire:
- Ks -- synonymous
- Kn -- non-synonymous
- Kn/Ks -- ratio of non-synonymous to synonymous changes. This is often used to detected neutral (Kn/Ks == 1), positive (Kn/Ks > 1), and purifying (Kn/Ks < 1) selection acting on a pair of genes.
Synonymous (Ks) and non-synonymous (Kn) site changes are calculated by:
- Performing a global sequence alignment of the protein sequences using the using the Needleman-Wunsch algorithm implemented in nwalign and written and maintained by Brent Pedersen using the BLOSOM62 scoring matrix.
- Back translating the protein sequence into aligned codons
- Using codeml of the PAML software package written and maintained by Ziheng Yang. We modified codeml for implementation in SynMap in order to minimize the number of read/write cycles to the hard drives, as well as allow it to be more easily run in parallel on multi-core servers. For each pairwise comparison of aligned codon sequences, codeml is run 5 times using its default parameter sets, and the lowest Ks is kept.
Running SynMap
Just press "Generate SynMap" when everything is configured. While SynMap runs, messages will be displayed as to the stage of the analysis that is being processed.
Interacting with results
Main results
- Cross-hairs help visually align syntenic region from different parts of the genome.
- Clicking on a chromosome panel in the initial results creates a zoomed-in panel.
Zoomed-in results
- Cross-hairs turn red when the mouse is over a gene pair that is a link to GEvo with that pair pre-loaded. GEvo can then be used to perform a high-resolution analysis of those genomic regions.
Example Results
Syntenic comparison of Arabidopsis thaliana and Arabidopsis lyrata
Maize and Sorghum
Strains of Escherichia coli K12
X-alignments of various bacterial genomes
Human-Chimp Whole Genome Comparison (Video)
FAQ
How do I sort the chromosomes by name instead of by size?
- Go to the "Display Options" tab
- select "Name" from the menu for "Sort Chromosomes by:"