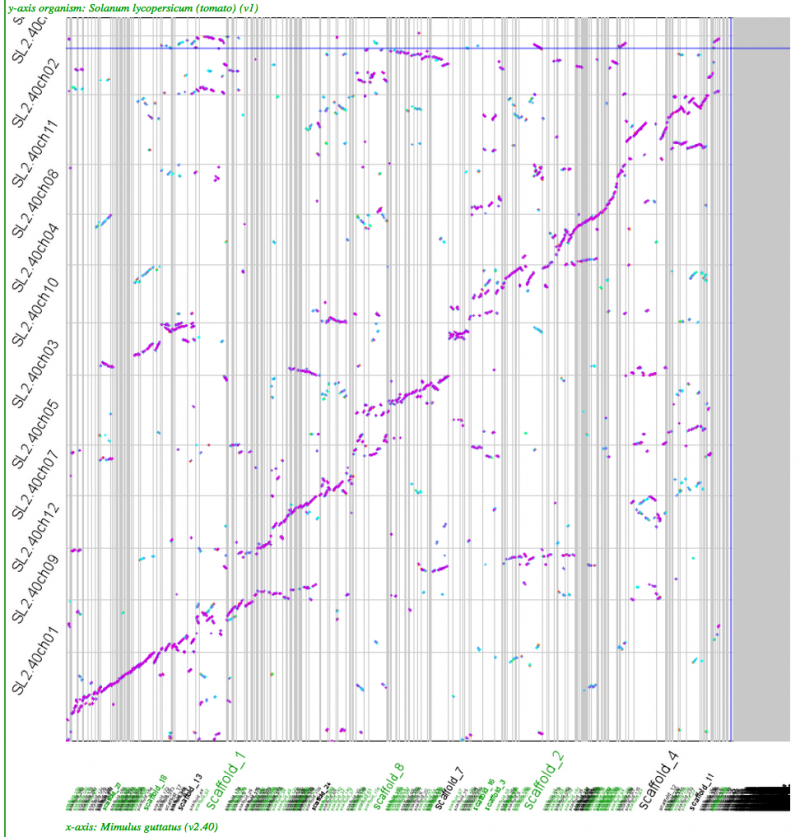
Mimulus v. tomato
Note: evaluation of tomato is difficult as its polyploidy is not a string tetraploidy or hexaploidy: Tomato genome
Whole genome syntenic dotplots
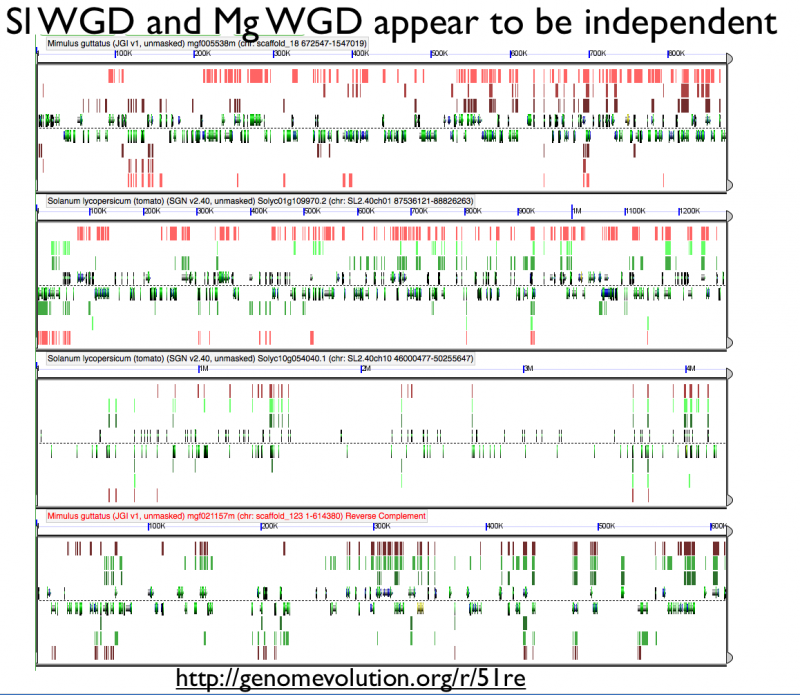
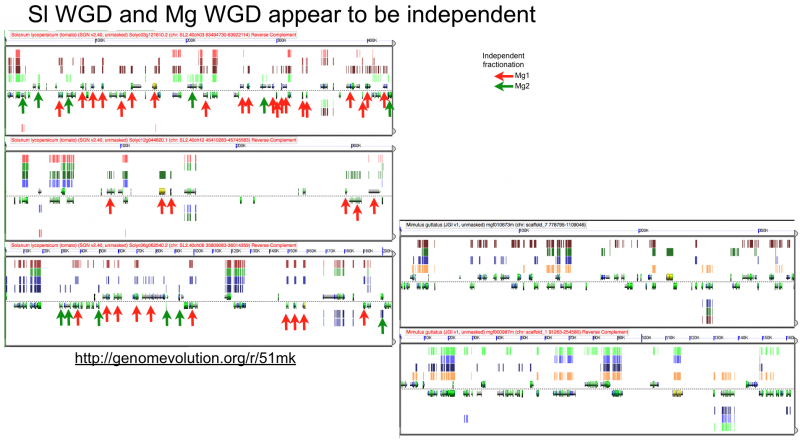
High-resolution analysis of syntenic regions with GEvo
Fractionation appears to be independent among intragenomic syntenic regions
Ks histograms of syntenic gene pairs
No syntenic depth screening
| Org 1 | Org 2 | Full Hist | Banded Hist | Notes |
|---|---|---|---|---|
| Mimulus guttatus | Solanum lycopersicum (tomato) | 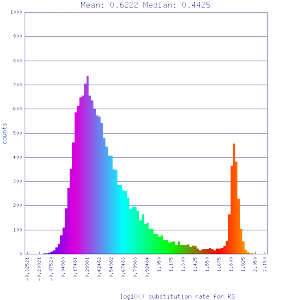 http://genomevolution.org/r/51li http://genomevolution.org/r/51li
|
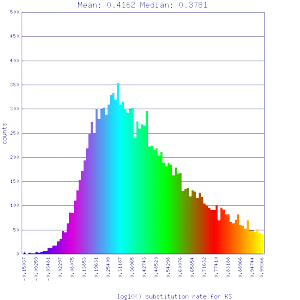 ]http://genomevolution.org/r/51up ]http://genomevolution.org/r/51up
|
|
| Solanum lycopersicum (tomato) | Solanum lycopersicum (tomato) | 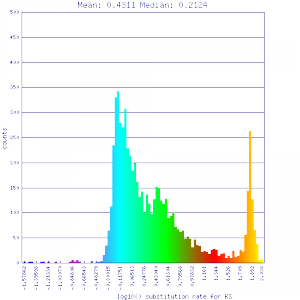 http://genomevolution.org/r/51uq http://genomevolution.org/r/51uq
|
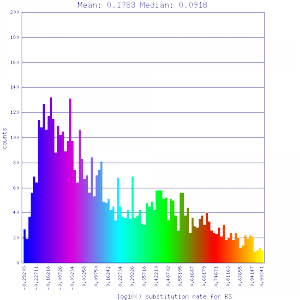 http://genomevolution.org/r/4tpr http://genomevolution.org/r/4tpr
|
|
| Mimulus guttatus | Mimulus guttatus | 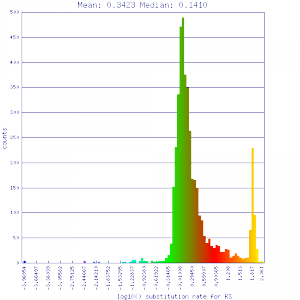 http://genomevolution.org/r/51v0 http://genomevolution.org/r/51v0
|
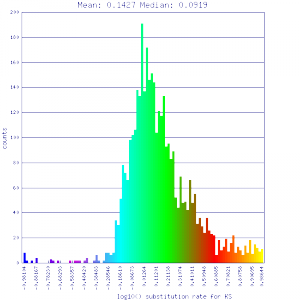 http://genomevolution.org/r/51v1 http://genomevolution.org/r/51v1
|
1:1 syntenic depth screening (identified primarily orthologous syntenic gene pairs
| Org 1 | Org 2 | Full Hist | Banded Hist | Notes |
|---|---|---|---|---|
| Mimulus guttatus | Solanum lycopersicum (tomato) | 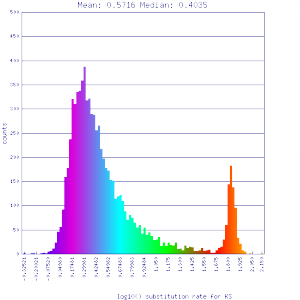 http://genomevolution.org/r/51uv http://genomevolution.org/r/51uv
|
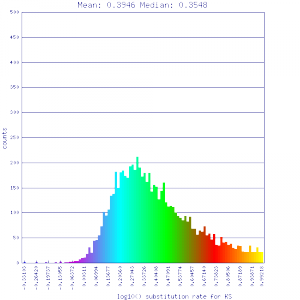 http://genomevolution.org/r/51uw http://genomevolution.org/r/51uw
|
Ortholog screening 1:1 Syntenic depth |
| Solanum lycopersicum (tomato) | Solanum lycopersicum (tomato) | 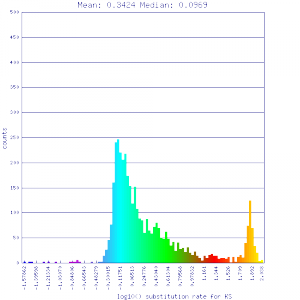 http://genomevolution.org/r/51v4 http://genomevolution.org/r/51v4
|
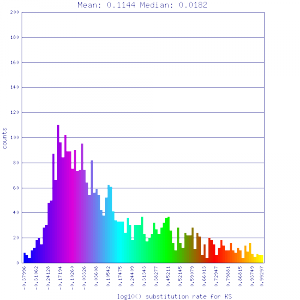 http://genomevolution.org/r/51v6 http://genomevolution.org/r/51v6
|
Ortholog screening 1:1 Syntenic depth |
| Mimulus guttatus | Mimulus guttatus | 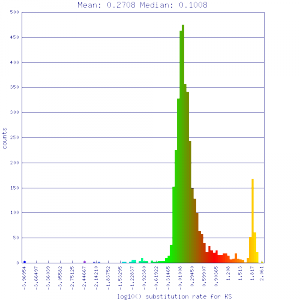 http://genomevolution.org/r/51v5 http://genomevolution.org/r/51v5
|
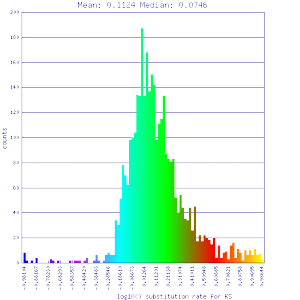 http://genomevolution.org/r/51v3 http://genomevolution.org/r/51v3
|
Ortholog screening 1:1 Syntenic depth |