Mycoplasma mycoides JCVI-syn1.0 Decoded
Background
Rumor has it that there is a code in one of the synthetic genomes by JCVI (passed along to me from Haibao Tang). Supposedly, this code contains an email address or a URL (or a secret message from Dr. V!). I wanted to know if CoGe's comparative genomics tools would make this relatively easy to do.
- WARNING: contains spoilers!
- Note: this puzzle is nearly 2 years old.
For those interested in doing the puzzle, this article has a good summary of the challenge:
- JCVI press release: http://www.jcvi.org/cms/press/press-releases/full-text/article/first-self-replicating-synthetic-bacterial-cell-constructed-by-j-craig-venter-institute-researcher/
- Article from Sigularity Hub on the secret message: http://singularityhub.com/2010/05/24/venters-newest-synthetic-bacteria-has-secret-messages-coded-in-its-dna/
And you will probably need the original article (and the Supplementary Data):
- Original Science Article on the genome: http://www.sciencemag.org/content/329/5987/52.abstract
Synthetic JCVI genomes in CoGe
- synthetic Mycoplasma genitalium strain JCVI-1.0: http://genomevolution.org/CoGe/OrganismView.pl?oid=35986
- synthetic Mycoplasma mycoides JCVI-syn1.0: http://genomevolution.org/CoGe/OrganismView.pl?oid=35385
Methods
- Find closest natural relatives
- Identify syntenic discontinuities (this is where the new JCVI code should reside
- Decode new sequence
- Identify coding scheme
- Probably using natural codon triplet encoding given that:
- 1x4 encoding = 4 letters
- 2x4 encoding = 16 letters
- 3x4 encoding = 64 letters
- Create cipher
- Using the genetic code: Given that there are 20ish natural amino acids, some of the codons will be appropriated for additional letters and symbols
- An example for students of expanded codon encoding (using neighboring codons for additional letters): http://nature.ca/genome/05/051/0511/0511_m205_e.cfm
- Using the genetic code: Given that there are 20ish natural amino acids, some of the codons will be appropriated for additional letters and symbols
- Probably using natural codon triplet encoding given that:
- Decode email address
- Identify coding scheme
- Validate email address
Closest natural relatives
| Syntenic dotplot of synthetic Mycoplasma genitalium strain JCVI-1.0 (y-axis) v. Mycoplasma genitalium strain G37 (x-axis) http://genomevolution.org/r/4mx1 | Syntenic dotplot of synthetic Mycoplasma mycoides JCVI-syn1.0 (y-axis) v. Mycoplasma mycoides subsp. capri strain GM12 (x-axis) http://genomevolution.org/r/4mx2 |
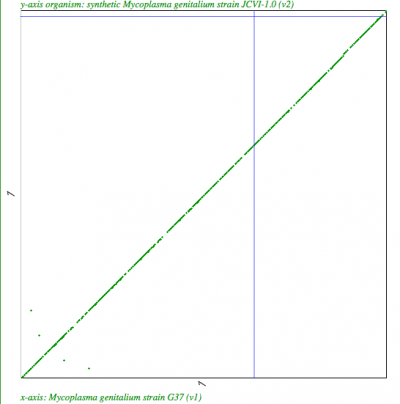
|
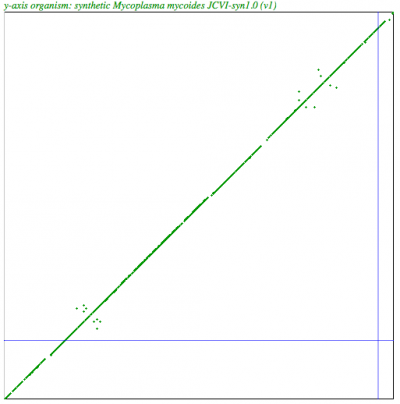
|
GEvo Analyses: high-resolution detection of syntenic discontinuities
Genitalium
Mycoides
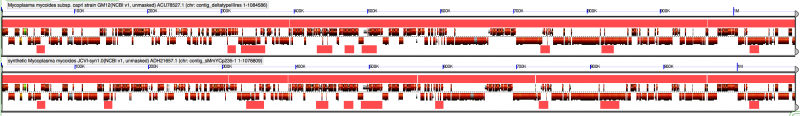

Identifying JCVI watermark sequences in mycoides using GEvo
No luck with first attempt at decoding
After searching through the watermarks, performing 6-frame translations on all of them, searching for "words" (e.g. CVI from JCVI), and looking at the darn sequences, and trying some funky stuff with overlayed 6-frame translations (in case the message was written in multiple reading frames simultaneously -- yes, highly unlikely!), I decided to dig into the literature a bit.
Example: Aligned 6-frame translation of watermark 4 shows nothing
Hints From the Paper and Craig V.
The paper describes four watermark sequences added to the genome, and these sequences were given in the supplementary information! Fortunately, using SynMap and GEvo, CoGe correctly identified these (along with the carryover piece of E. coli), but getting them from the literature would have been a lot easier. There were additional hints as to the information in the watermarks. Specifically that three of the four watermarks have quotes (given) and that one contained the secret message. There is also a Skype interview with Craig that is fun to watch.
- Paper: http://www.sciencemag.org/content/329/5987/52.abstract
- Sup Data: http://www.sciencemag.org/content/suppl/2010/05/18/science.1190719.DC1/Gibson.SOM.pdf
- http://www.popsci.com/science/article/2010-05/venter-institutes-synthetic-cell-genome-contains-hidden-messages-watermarks
- http://en.wikipedia.org/wiki/Mycoplasma_laboratorium
- JCVI code phrases
- JCVI watermarks (without end sequences)
Decoding part one
The code is something different than using protein translation. Also, there needs to be a way to code special characters, such as "," or ":", etc.
JCVI GEvo analysis of watermarks
First success at decoding:
The fourth phrase:
What I cannot build, I cannot understand
Contains a direct repeat sequence. Let's see if this exists in the 4th watermark sequence.
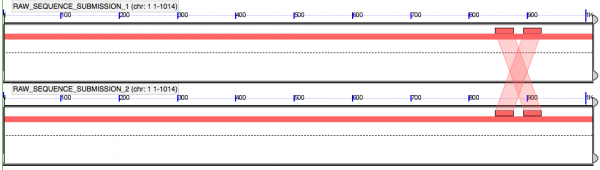
- Direct Repeat Sequence (DRS): 100% match, 30 characters
ATACTGATATTTTAGTGCTGCCGTTGAATA
- Interesting. DRS is 30 characters "cannot" is 6 == looks like 5 nucleotide encoding scheme. However, there doesn't appear to be enough space between the two words...
- Almost: it is " I cannot " -- 10 characters! coding is 3 nucleotides. Where have I seen that before?
Cracking the code (from the repeat sequences)
ATA AAC CTG GGC TAA <s> l i f e TGA ATA TAG GCT ATA TGA TCA TAA CAT ATA t <s> a s <s> t h e y <s> ATA CTG ATA TTT TAG TGC TGC CGT TGA ATA <s> I <s> c a n n o t <s> :)
- Notes:
- spaces all use the same triplet (ATA)
- lower case "i" and upper-case "I" both use the same triplet (CTG): code is case insensitive
Decoding: First pass
This part requires writing a program to do the decoding. The overall logic isn't too hard.
- Read in the watermark sequences
- Read in the quotes
- Use the decoding sections (from the repeat sequences above) to associate the correct quote with a watermark sequence
- Identify the location of the decoded section in both
- Map the remaining part of the quote (up and downstream of the matches) to the watermark sequences
- Extract those sequences and quotes
- Use the decoded quotes to build a cipher
- Use the cipher to decode as much of the watermarks as possible.
Decoded: First pass
Not too bad.
There is an obvious "decoding" section in the first watermark (along with some HTML tags)
Second pass: 95% solved (good enough for computational biology)
Solving the code from this point is pretty straight forward. Mostly a matter of looking for obvious characters, plugging them in, rerunning the program, and repeat. There is a decoding section in the first watermark, but I did not decode all of the symbols. I'm guessing there are a variety of other ASCII characters such as "(){}[]|?/\" and whatnot. "?" in the text are those symbols that I did not decode.
Watermark one
J. CRAIG VENTER INSTITUTE 2009 ABCDEFGHIJKLMNOPQRSTUVWXYZ 0123456789?@??-??=/:<?>??????"??!'., SYNTHETIC GENOMICS, INC. <!DOCTYPE HTML><HTML><HEAD><TITLE>GENOME TEAM</TITLE></HEAD><BODY><A HREF="HTTP://WWW.JCVI.ORG/">THE JCVI</A><P>PROVE YOU'VE DECODED THIS WATERMARK BY EMAILING US <A HREF="MAILTO:XXXXXXXX@JCVI.ORG">HERE!</A></P></BODY></HTML>
Watermark two
MIKKEL ALGIRE, MICHAEL MONTAGUE, SANJAY VASHEE, CAROLE LARTIGUE, CHUCK MERRYMAN, NINA ALPEROVICH, NACYRA ASSAD-GARCIA, GWYN BENDERS, RAY-YUAN CHUANG, EVGENIA DENISOVA, DANIEL GIBSON, JOHN GLASS, ZHI-QING QI. "TO LIVE, TO ERR, TO FALL, TO TRIUMPH, TO RECREATE LIFE OUT OF LIFE." - JAMES JOYCE
Watermark three
CLYDE HUTCHISON, ADRIANA JIGA, RADHA KRISHNAKUMAR, JAN MOY, MONZIA MOODIE, MARVIN FRAZIER, HOLLY BADEN-TILSON, JASON MITCHELL, DANA BUSAM, JUSTIN JOHNSON, LAKSHMI DEVI VISWANATHAN, JESSICA HOSTETLER, ROBERT FRIEDMAN, VLADIMIR NOSKOV, JAYSHREE ZAVERI. "SEE THINGS NOT AS THEY ARE, BUT AS THEY MIGHT BE."
Watermark four
CYNTHIA ANDREWS-PFANNKOCH, QUANG PHAN, LI MA, HAMILTON SMITH, ADI RAMON, CHRISTIAN TAGWERKER, J CRAIG VENTER, EULA WILTURNER, LEI YOUNG, SHIBU YOOSEPH, PRABHA IYER, TIM STOCKWELL, DIANA RADUNE, BRIDGET SZCZYPINSKI, SCOTT DURKIN, NADIA FEDOROVA, JAVIER QUINONES, HANNA TEKLEAB. "WHAT I CANNOT BUILD, I CANNOT UNDERSTAND." - RICHARD FEYNMAN
Conclusion
CoGe definitely helped. Using CoGe did permit the rapid identification of the watermark sequences and checking whether they were unique sequences (e.g. came from E. coli). However, those watermark sequences were given in the supplementary data. Having some of the code broken made the cracking of it pretty simple when using GEvo to compare within and among the watermark sequences. This quickly showed that watermark one was very different in structure than the other three. In addition, each of the other three watermark sequences has a relatively large identical repeat sequence, which permitted decoding the first set of works, locating their placement within the quotations and the watermarks, and using them to build a cipher to decode the rest of the sequences. All in all, a lot more fun than Sudoku!
Of course, it seems to take quite a while for new synthetic genome puzzles to come out. . .
Confirmation from JCVI: 83rd person to crack the code
After emailing the special address last night, I received a response this morning. After nearly two years, less than 100 people have solved this puzzle. Still, even with CoGe, I wouldn't have been able to come in around 3h.
From: "Montague, Michael" <MMontague@jcvi.org> Subject: RE: A bit late Date: March 23, 2012 7:53:28 AM MST To: Eric Lyons <elyons.uoa@gmail.com> Hi Eric, You are the 83rd person or group to decode the watermarks. The first decoder was a recently graduated student from U. Penn. named Andrew Ettenger only 3 hours and 13 minutes after the watermark sequences were released. --Mike Montague From: Eric Lyons [mailto:elyons.uoa@gmail.com] Sent: Thursday, March 22, 2012 11:55 PM To: XXXXXXXXXXXXX Subject: A bit late To JCVI Team, I know this is quite a bit late, but I finally heard about your secret code challenge. I was able to use the CoGe Comparative Genomics system to help with various parts of it. Overall, a lot of fun! If you are interested, here are my notes from cracking this puzzle: genomevolution.org/wiki/index.php/Jcvi_code Hope you make more of these soon! Thanks, -eric --------------------------------------------------- Eric Lyons, Ph.D. Comparative Genomics
Additional thoughts
As synthetic DNA technology improves, the opportunity will come where anyone can create DNA sequences of their choosing; as with all technology, there are great opportunities and great concerns. The ethical implications are quite important and are still an actively discussed and debated. The only area on which I feel any competence to comment is the importance of tools to quickly analyze DNA sequences. While the exercise (or game) presented here was quite fun, it does show that tools such as CoGe are needed to quickly permit the analysis and decoding of genomic data. In case harmful synthetic organisms are created, the methods presented here should permit the rapid deconstruction of their genomes in order to:
- identify which parts are from which organisms
- identify which parts are novel
- identify information contained in those parts