Non-X-alignments
Jump to navigation
Jump to search
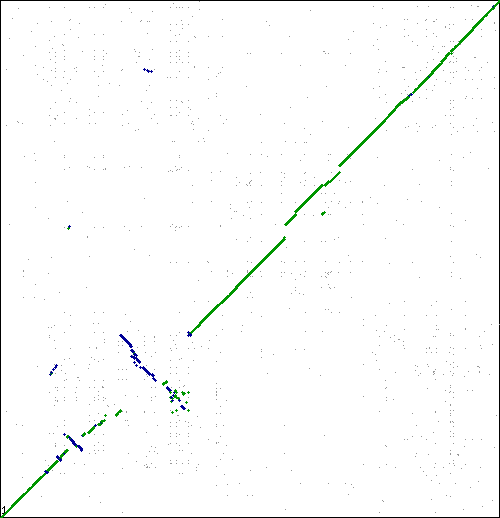
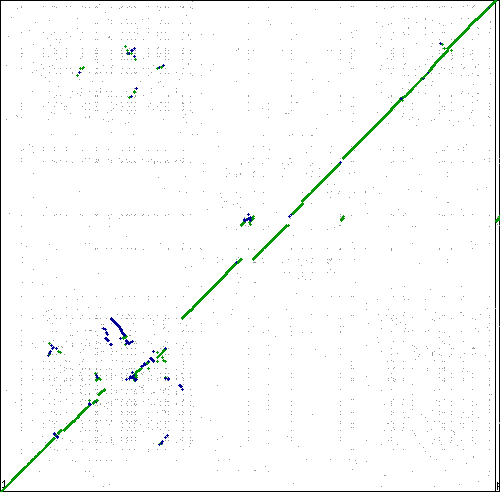
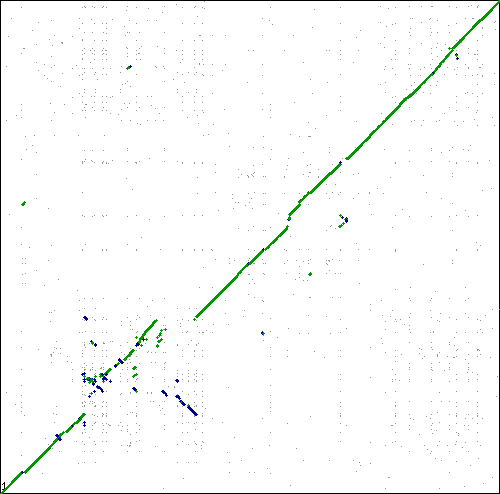
This tracks examples of bacteria and archaea syntenic dotplots which do not show the classical x-alignment pattern of inversions. One possible methodological reason for these non-x-alignments is mis-assembly.
Genome-wide random orientation


Local region of syntenic instability
These are interesting. Lots of inversions at the same local sub-regions, but not around a single point in the genome. Multiple events appear to be independent by comparison to different genomes. Perhaps there is some factor contributing to a local region of syntenic instability?