Phylogenetics in CoGe
Find a sequence of interest.

To begin, you'll need at least one sequence:
- You can use CoGe's tools such as FeatView or GenomeView
- Or, get a sequence from somewhere else such as NCBI
If you use CoGe to get a sequence, you can send it directly to CoGeBlast to find homologs.
Search for potential homologs in other genomes using CoGeBlast
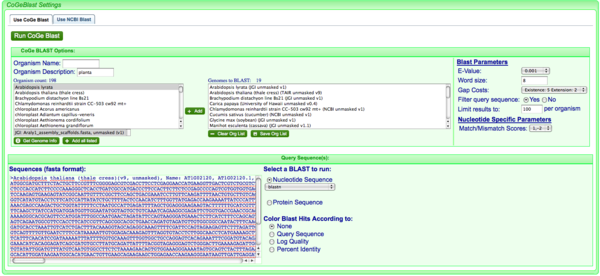
CoGeBlast lets you search your sequence(s) against any number of genomes in CoGe. In this case, organisms were identified that have "planta" in the organism description, which will find all plant genomes in CoGe. CoGe's organism descriptions usually follow NCBI's taxonomic description:
Eukaryota;Viridiplantae;Streptophyta;Embryophyta;Tracheophyta;Spermatophyta;Magnoliophyta;eudicotyledons;core eudicotyledons;rosids;eurosids II;Brassicales;Brassicaceae;Arabidopsis;
and a search for "planta" matches anything with "Viridiplantae".
Evaluate potential homologs in CoGeBlast and select the "good" ones

After selecting and searching genomes in CoGeBlast, its visualization tools make it easy to identify hits that match homologs. By clicking on an HSP, a popup graphic will be displayed showing the regions matched between the query sequence and a genome. The blast hit you clicked on in colored yellow, and all other blast hits between that query sequence at the genomic region are colored red. This makes it easy to see that the query sequence has full coverage in a genomic region, even if there are several blast hits between those sequences.
Send the identified homologs to FeatView
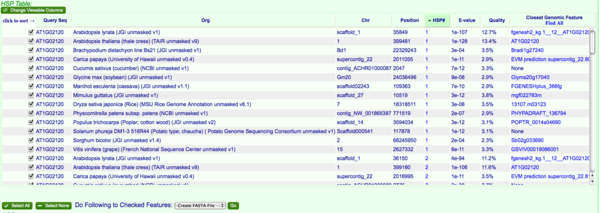
CoGe's blast will identify genomic features overlapping with a blast hit. This makes it so that you can check the sequence and mark it to be sent to other tools in CoGe. In this case, we are going to send all overlapping genomic features to FastaView. Don't worry if you check the of the same overlapping sequence more than once, all duplicate submissions will be collapsed to a single entry.
Send the selected homologs to FastaView

FastaView is CoGe's tool for viewing many fasta sequences. It has a button you can press to translate DNA sequence to protein sequence.
Modify sequences in FastaView
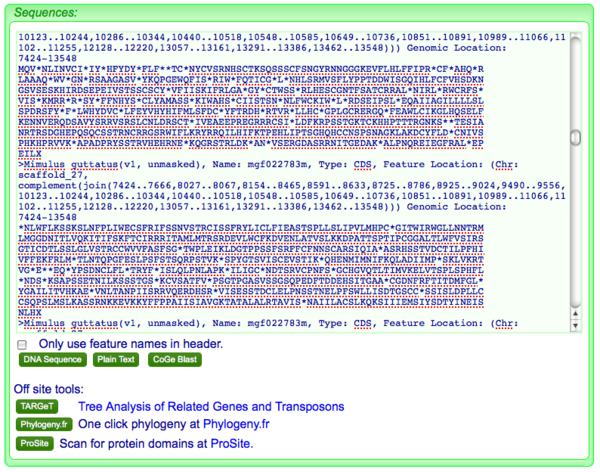
FastaView will try to identify the correct reading frame for DNA sequence by identifying a start methionine (M), a stop codong (*) and to intragenic stop codons. If it can't meet these three criteria, it will return all 6 reading frame translations. If this happens, as is shown here, you will need to remove any translation that is not correct. You can do this by highlighting the bad fasta sequence in the text-box and pressing the delete key.
Likewise, if you have additional sequences you'd like to add, just copy and paste them into the text-box in fasta format. Remember to add back your original sequence if you need to!
To send your sequences to phylogeny.fr for phylogenetic anlaysis, just press the "phylogeny.fr" button. All the sequences in the text-box will automatically be submitted and the analysis at phylogeny.fr automatically started.
Send sequences to phylogeny.fr




Phylogeny.fr's pipeline consists of:
- Multiple sequence alignment using MUSCLE
- Phylogenetic tree reconstruction using PhyML
- Phylogenetic tree visualization using TreeDyn
Video Tutorial
This short video walks through the example detailed above.