SynMap3D
Note: This is the documentation for SynMap3D, if you are looking for SynMap2 look here.
Contents
Background
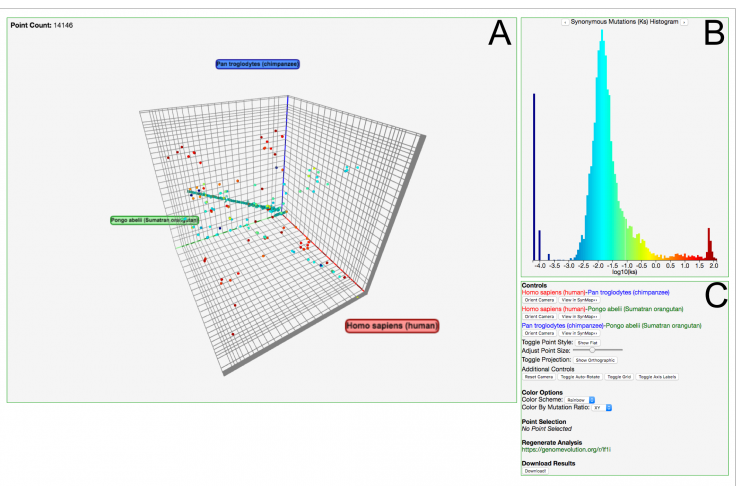
SynMap3D is a web-based whole-genome synteny browser that both identifies and visualizes regions of synteny (common ancestry) between three genomes. Results are displayed as a three dimensional dotplot (scatter plot), with each axis representing a linearized version of the genome. Alternating light/dark axis colors and grey grid lines distinguish between different chromosomes. Each point represents a syntenic gene pair shared between all three genomes.
Getting Started
Launching a SynMap3D Analysis
Launching SynMap3D is accomplished using the familiar CoGe "wizard" which guides through each step of selecting organisms, chosing options, reviewing selections then launching calculations and visualization.
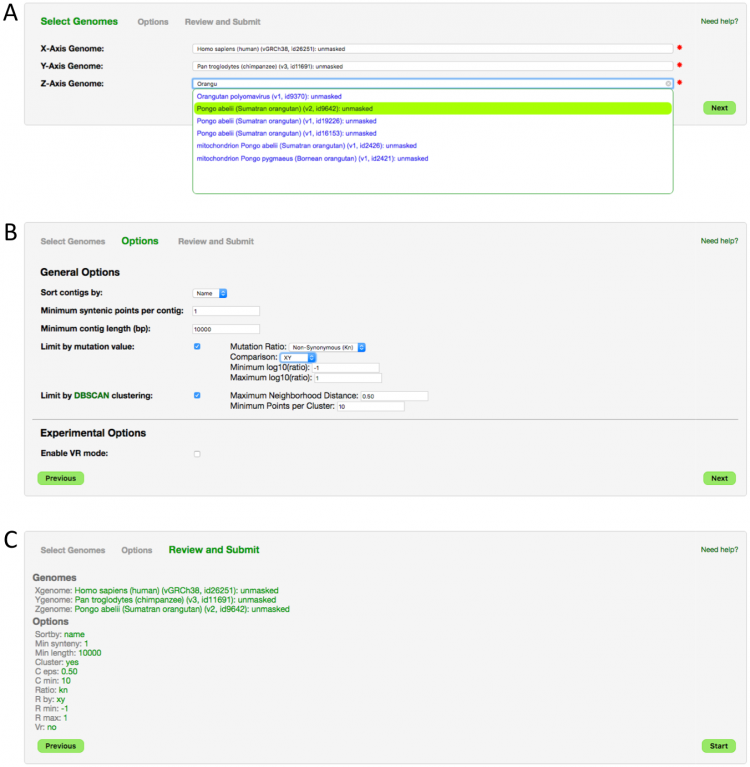
Step 1: Select Genomes (Figure 1A)
Three genomes must be selected for a SynMap3D analysis. Start typing an organism name or a genome ID and the search will find matching genomes. After three genomes are selected, choose 'Next'.
Step 2: Choose Options (Figure 1B)
Select all desired options:
- General Options
- Sort Contigs By: Use this option to adjust how chromosomes/scaffolds/contigs are sorted during the "linearization" of genomes (conversion to graph axes). 'Name' will use a natural sorting algorithm to order them logically by name (i.e. 1, 2A, 2B, 3, 4). "Length" will order by length of each contig (largest to smallest).
- Minimum syntenic points per contig: Use this option to set a minimum threshold of syntenic points per chromosome/scaffold/contig for display. For example, a setting of 10 means that if a contig has less than 10 syntenic points, it will not be displayed. This is very useful for incompletely assembled genomes and those with many pieces.
- Minimum contig length (bp): This sets the minimum chromosome/scaffold/contig length (in basepairs) for display. For example, a setting of 10,000 will remove any pieces less than 10kb. Like above, this is very useful for genomes with many small pieces.
- Limit by mutation value: This option allows you to set minimum/maximum Kn, Ks, or Kn/Ks values for display. This is particularly useful if you are dealing with polyploid genomes with large amounts of synteny and are only interested in those points with certain evolutionary characteristics.
- Mutation Ratio: Use this option to select the mutation value you would like to limit by (Kn, Ks or Kn/Ks).
- Comparison: Use this option to select which pairwise or three-way calculation you would like to use to limit by.
- Minimum log10(ratio): This is the minimum log10 transformed mutation value to consider.
- Maximum lot10(ratio) This is the maximum, lot10 transformed mutation value to consider.
- Limit by DBSCAN clustering: This option allows for removing of syntenic points that do not fall into larger syntenic regions. The DBSCAN algorithm is used for this filtering. For details about the specific implementation and parameters, see sklearn.
- Maximum neighborhood distance: The maximum distance between two points for them to be consitered within the same neighborhood.
- Minimum points per cluster: The minimum number of additional points that must fall within a points neighborhood to retain that point.
- Experimental Options
- Enable VR mode: Select this option to enable the VR visualizer. Note, this requires a VR headset and compatible web browser (see below). If you do not meet these requirements, the visualization will load with the normal renderer and a message will be displayed with the error.
Step 3: Review Options (Figure 1C)
The final wizard step is reviewing your selections. If everything looks good here, go ahead and "Launch". Otherwise, use the "Previous" button to navigate back and adjust accordingly.
- Zoom
- Scroll (wheel or touch pad).
- Rotate
- Click and drag.
- Shift
- Arrow keys.
- Select Point
- Click on point, selected point will turn black. (NOTE: If having trouble selecting a desired point, try moving around to get a different angle or adjusting zoom)
- View Specific Comparison
- Use "Orient Camera" option for desired comparison(Figure 2C).
- Reset camera
- Use "Reset Camera" under "Additional Controls" (Figure 2C).
- Automatically Rotate around Dotplot
- Select "Toggle Auto-Rotate" from "Additional Controls" (Figure 2C).
- Hide Grid
- Select "Toggle Grid" from Additional Controls (Figure 2C).
- Hide Axis Labels
- Select "Toggle Labels" from Additional Controls (Figure 2C). This will remove the 3D labels and place them in 2D on the lower part of the rendering.
- Enter VR mode
- VR mode must be enabled from options, then click goggle logo in lower right corner.
- Alternate between Kn, Ks, Kn/Ks Histograms
- Use arrows on either side of histogram title.
- Select Subset
- Click and drag ('brush') across histogram.
- Move Selection
- Click and drag existing selector box.
- Adjust Selection
- Use handles on either side of selection box.
- Reset to Full Dataset
- Click in any unselected region.
Additional Control (Figure 2C)
- Alternate Projection (Ortographic vs Projection)
- Use "Toggle Projection" button.
- Change Point Coloring Scheme
- Use "Color Scheme" drop down under "Color Options
- Increase/Decrease Point Size
- Use "Adjust Point Size" slider.
- Toggle Between Round Points & Flat Points
- Use 'Toggle Point Style' button.
- Apply Color Scheme to Differentiate Between Peaks
- Select "Auto" from Color Scheme drop down, then adjust slider until sensitivity is as needed.
- Change Mutation Value Representation
- Use "Color By Mutation Value" drop down.
- View Microsynteny Analysis of a Point
- Select point (click) then choose "Compare in GEvo" under "Point Selection"
- Compare two genomes in SynMap2
- Select "View in SynMap" for desired comparison.
- Download Raw Data
- Use "Download" button in "Download Results".
Examples
| Species 1 | Species 2 | Species 3 | Special Options | Link |
|---|---|---|---|---|
| H. sapiens (human) | P. troglodytes (chimpanzee) | P. abelii (orangutan) | Defaults | https://genomevolution.org/r/lf1i |
| H. sapiens (human) | P. troglodytes (chimpanzee) | P. abelii (orangutan) | Outlier points (non-clustered) removed. | https://genomevolution.org/r/lfbe |
| H. sapiens (human) | M. musculus (mouse) | C. familiaris (dog) | Defaults | https://genomevolution.org/r/lfbg |
| A. thaliana | A. lyrata | B. rapa | Limit A. thaliana/B.rapa log10(Ks) values < 0.8, remove outliers. | https://genomevolution.org/r/lfbn |
| Maize | Sorghum | Foxtail Millet | Remove all but very large clusters. | https://genomevolution.org/r/lg40 |
| O. niloticus (Nile tilapia) | S. salar (atlantic salmon) | D. rerio (zebrafish) | Defaults | https://genomevolution.org/r/mmuf |
| E. coli (MG1655) | E. coli (DH10B) | E. coli (NCM4299) | Defaults | https://genomevolution.org/r/lfbh |
Linking to SynMap3D
All options can be pre-filled using a special-format link to SynMap3D.
Base URL: https://genomevolution.org/coge/SynMap3D.pl?
Options:
- NOTE: Separate multiple options with a semicolon (;).
- X-Axis Genome: x_gid=<genomeID>
- Y-Axis Genome: y_gid=<genomeID>
- Z-Axis Genome: z_gid=<genomeID>
- Sort Contigs By: sort=<method=name/length>
- Minimum Syntenic Points per Contig: min_syn=<minimium syntenic points>
- Minimium Contig Length: min_len=<length in basepairs>
- Limit By Mutation Value: ratio=<values=kn/ks/knks>,<comparison=xy/xz/yz/mean/median>,<minimum>,<maximum>
- Limit By DBSCAN Clustering: cluster=<neighborhood distance>,<points per cluster>
- Enable VR Mode: vr=<0/1>
FAQ
- I want to get a good view of just a two way comparison, is there a way to make the visualization more clear?
- Definitely! Use "Toggle Projection" to "Show Orthographic". This projection removes perspective and gives a very clear two-way view. For more information on ortographic vs perspective projection, see this great illustration
- It's difficult for me to resolve colors between close points, they just look grey!
- This phenomenon is caused by the spherical shape of the points when only the darkest shoulder can be seen. You can do three things to get around this. First, try rotating the scene a bit, this can usually clear it up by getting a different angle. Second, you can toggle point style to "Flat". Third, you can increase the size of points to make the overlap less significant.
- How can I use VR-mode?
- VR-mode requires both a VR-compatible web browser and a VR headset. For information on setting up a VR-compatible browser, please visit https://webvr.info/
- What web-visualization technologies are used in SynMap3D?
- The three-dimensional dot plot is rendered with WebGL using Three.js. Histograms are pure D3.
- How many points can SynMap3D support?
- This really depends on the individual user's system. By default, we enforce a 100,000-point limit to ensure a smooth experience for all users. Raw data from analyses with over 100K hits is still available for download, but the renderer will be replaced by a warning message. If you need to visualize more than 100K points and believe your system can handle it, contact CoGe support for a temporary increase in the point count limit.
- How can I reduce the number of points displayed?
- There are two steps at which point number can be reduced: pre-visualization (through the wizard options) or post-render (by selecting histogram regions). During the wizard step, the four general options "Minimum syntenic points per contig", "Minimum contig length (bp)", "Limit by mutation value", and "Limit by DBSCAN clustering" can all be used to reduce point count (see 'Getting Started' for detailed information about each option. After rendering, data can be subsetted by selecting a region of the histogram. The visualization will live-update to display only the points falling within those values.
- I'm having trouble selecting the point I want, do you have any tips?
- If you are having trouble selecting a point, it frequently helps to slightly move the visualization around a bit. Some of the calculations can get confused with overlapping points or certain views, and slightly altering the view can help adjust calculations and get the correct point selected.
- Is SynMap3D open source?
- Absolutely! All CoGe tools are open source and can be viewed at our team's GitHub repository (https://github.com/LyonsLab/coge). If you are specifically interested in SynMap3D, most of the visualizations are controlled by the javascript files in web/js/syn3d.
- Question not answered here?
- Feel free to contact Asher @ ahaug (at) email (dot) arizona (dot) edu