New to CoGe

Contents
Getting CoGe to run
You just need a web browser and a connection to the internet to start working with CoGe: Here are the details.
Get a user account
While you can use CoGe anonymously, getting a user account lets you
- Have a user profile page
- Store and share private data
- Create Notebooks which are collections of data (genomes, experiments)
- Save your analytical history so you can easily organize and find previously run analyses
CoGe's design
CoGe is designed to help make access to genomic information easy and facilitate doing complex comparative genomics analyses. The web-based tools give access to thousands of genomes, allowing you to find what you want and compare it to what you. Each tool in CoGe is designed for one purpose, and is linked to other tools that in turn do something specific creating an Open-ended analysis network. This approach means that you can start an analysis from many different places with different types of genomic data (e.g. a sequence or an organism name), and there is no defined end to an analysis. You have tools to explore different aspects of genomic biology and you stop when you get your answer.
How to start with CoGe
There are several places to begin an analysis depending on what your question is and what data you have in hand. The tools listed on CoGe's front page are often used to begin an analysis. Each tool has a brief description of what it does. For example, if you have a sequence and you want to find it in a set of genomes, begin with CoGeBlast. If you have an organism in which you are interest, begin with OrganismView. If you are interested in comparing two genomes, begin with SynMap
If you are interested in learning about what kind of questions you might answer using CoGe, you can begin reading some tutorials. However, know that just because your particular question is not listed there doesn't mean it is not possible to answer it with CoGe. CoGe's tools are designed to let you explore your ideas as easily as possible, and not force you to adapt your ideas to its tools. If you have a question about getting started, feel free to contact us.
Just want to play with a genome
If you just want to play with a genome and start with some comparative genomics, begin with OrganismView. Just type in part of the name of an organism that interests you in the name box and see if there is a genome for it. If you want a suggestion, start with your good friend Escherichia coli, the common human gut commensal.
You can begin exploring its genome by clicking on:
- "Launch genome viewer" in the lower left of the web page to browse its genome
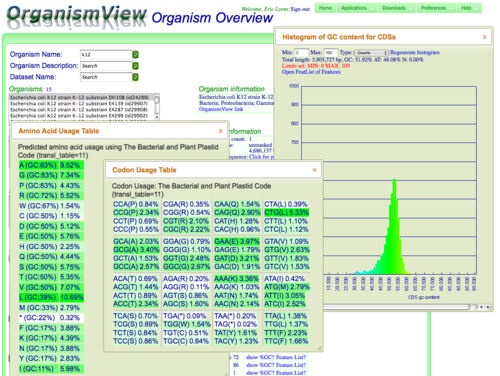
- "Click for features" under the Genome Information box to get an overview of the annotated features in its genome. If there are CDS (protein coding features) in the genome, you can generate a histogram of GC content of protein coding genes, a codon usage table, and an amino acid usage table.
To do some comparative genomics, grab a genomic region from the genome browser (GenomeView), find putative syntenic regions in other E. coli genomes using CoGeBlast, and compare those regions to look for synteny using GEvo.