GEvo
Contents
- 1 Introduction
- 2 Getting started
- 3 Sequence Submission
- 4 Alignment Algorithms
- 5 Results
- 6 Regenerating/Saving a GEvo Analysis
- 7 Modifying result graphics
- 8 Merging Analyses
- 9 Refining an analysis
- 10 Hints and Tricks
- 11 Example Analyses
- 12 Linking to GEvo
- 13 Exporting Images for Publication
- 14 Tutorials
- 15 References
Introduction
The purpose of GEvo is to compare multiple genomic regions from any number of organisms using a variety of different sequence comparison algorithms in order to quickly identify patterns of genome evolution
Getting started
- Select genomic regions to analyze
- Select a sequence alignment algorithm appropriate for the sequences and questions in mind
- Press "Run GEvo Analysis!" button
To alternate between areas to configure an analysis, select the appropriate tab.
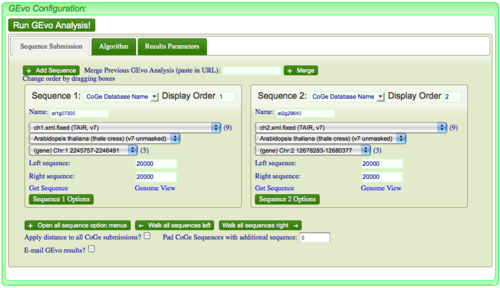
Sequence Submission
Select the "Sequence Submission" tab to open these options. Here, you can specify sequence submission boxes for each sequence that will be submitted for a GEvo anlaysis. This is also were you can adjust the amount of sequence analyzed, select which sequences are analyzed, reverse complement a sequence, mask a sequence according the the genomic features it contains, and change the display order of sequences.
Adding a sequence
To add another sequence submission box, press the "Add sequence" button. After pressing, a new sequence submission box will appear
Select the type of sequence
There are three types of ways that you can submit a sequence to GEvo:
- Using a CoGe genomic feature name
- Specify a GenBank Accession for automatic retrieval from NCBI
- Paste a sequence in Fasta or GenBank format
You can select the sequence submission type from a drop-down menu located next to the name header in a sequence submission box. E.g.: Sequence 1.
Specifying the amount of sequence analyzed
Each sequence allows you to specify the amount of sequence to be analyzed.
- For sequences retrieved from CoGe's database, you can specify the amount of sequence to the left and right of the genomic feature you specified by name.
- For sequences retrieved from NCBI, you specify a start position and the length of sequence from that position. If the length is left blank, the entire sequence from the specified start position to the end of the sequence is used.
- For direct sequence submissions, you specify a start position and the length of the sequence from that position. If the length is left blank, the entire sequence from the specific start position to the end of the sequence is used.
If you want to change all the positions of CoGe submissions at once (e.g. add 1000000 nt left and right of all sequences), just type in the amount of sequence into the box next to "Apply distance to all CoGe submissions".
Likewise, you can do something similar with the box next to "Pad CoGe Sequences with additional sequence". However, when the results are returned, the position slider bars are positioned based on the amount specified, not taking into account the amount added through padding.
Skip a sequence
- You can access this option by opening the "sequence options" menu by click on the button located at the bottom of the sequence submission box (e.g. Sequence 1 Options).
After a preliminary GEvo anlaysis, you may want to drop a sequence from the analysis. For example, if the region is repeated in another submission or if there the pattern you were seeking is not present (e.g. synteny). To skip a sequence, just select "yes" from the row "Skip Sequence" in the options shown at the bottom of each sequence submission box.
Making a sequence a "reference sequence"
- You can access this option by opening the "sequence options" menu by click on the button located at the bottom of the sequence submission box (e.g. Sequence 1 Options).
A reference sequence is one to which all other sequences are compared. By default, all sequences are selected as reference sequences. However, you may wish to only specify a couple of reference sequences. For example, you are searching for syntenic regions from genome B to genomic region A. By turning off sequences as reference sequences, the number of pair-wise comparisons is minimized so the analysis is processed more quickly. Also, this can help simplify visualizing and interacting with the results. Especially if one genome has lots of repetitive sequences.
Reverse complement a sequence
- You can access this option by opening the "sequence options" menu by click on the button located at the bottom of the sequence submission box (e.g. Sequence 1 Options).
After a preliminary GEvo analysis, you may want to "flip" a sequence. To do this, select "yes" for the row "Reverse complement" located at the bottom of the sequence submission boxes.
Masking a sequence
- You can access this option by opening the "sequence options" menu by click on the button located at the bottom of the sequence submission box (e.g. Sequence 1 Options).
This option allows you to mask a sequence based on its annotated genomic features:
- CDS: Mask all protein coding sequences
- All RNA: Mask anything which codes for RNA including rRNA, mRNA, tRNA, siRNA
- Non-CDS: Mask everything except protein coding sequences (very useful when identifying synteny)
- Non-genic: Mask everything that is not annotated as a gene.
Changing the display order of sequences
Sequences are displayed according to the order of the sequence submission boxes. To change this order, just click and drag the title of a sequence submission box into a new position.
Alignment Algorithms
Current GEvo can use:
- BlastZ: DNA-DNA Local Alignment Algorithm. Good for finding large regions of conserved sequence.
- BlastN: DNA-DNA Local Alignment Algorithm. Good for finding small regions of conserved sequence.
- TBlastX: Translated DNA-Translated DNA Local Alignment Algorithm. Good for finding small regions of divergent, but evolutionarily conserved, genomic sequence where protein translated sequence is more conserved than DNA sequence.
- Chaos: DNA-DNA Local Alignment Algorithm. Good for finding small regions of conserved sequence. Uses fuzzy matches so it can seed its alignment on small sequences than BlastN. However, it is slower than BlastN.
- DiAlign: DNA-DNA Global Alignment Algorithm. Global alignment can be seeded using local alignment algorithm. Good for alignment the entire sequence.
GEvo supports using Chaos, BlastN, and BlastZ for seeding DiAlign.
- Lagan: DNA-DNA Glocal Alignment. Using a hybrid alignment approach.
Picking an algorithm to use: blastn versus blastz
Of these algorithms, blastn and blastz are the two that are most useful for most people. These two algorithms are local alignment algorithms and will find regions of sequence similarity located anywhere between two sequences. The primary difference of these algorithms in terms of their utility are:
- blastn finds small regions of sequence similarity
- blastz finds large regions of sequence similarity by combining smaller regions of sequence similarlity
Use blastz if you are comparing large genomic regions and/or looking for synteny by identifying collinear series of genes Use blastn if you are comparing gene models, analyzing exons, searching for CNSs], or looking for conserved motifs
Results
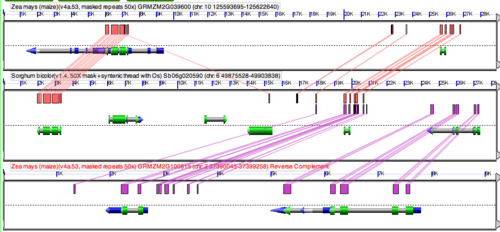
GEvo's results are displayed in an interactive system called gobe that lets you connect regions of similar sequence, and get additional information about genomic features. Please follow this link for more information about gobe.
Each panel represents a genomic region, with the dashed line in the middle separating the top and bottom strands of the chromosome. If gene models are present, they are drawn as composite colored arrows above and below this line if they are read from the top and bottom strand respectively. Usually, the full gene is the gray arrow, on top of which is the mRNA (blue), on top of which is protein coding sequence (CDS). There are other colors and icons that represent other types of genomic features that are described [[GenomeView_examples | here]. Above and below the gene models will be the identified regions of sequence similarity. These are represented by colored boxes. The location of the colored box above or below the dashed line signifies of whether the match is in the (++) or (+-) orientation respectively. Each pairwise comparison will have its regions of sequence similarity drawn in a separate track (both above and below the dashed line) and are usually different colors from one another (though that is configurable). To see which region matches which other region, just click on a colored box, and a transparent wedge will be drawn connecting it to its partner region. For more information about GEvo's interface, see the documentation on gobe.
Regenerating/Saving a GEvo Analysis
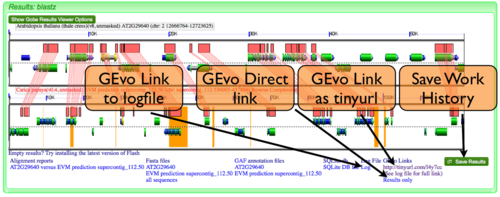
GEvo Links
After results are generated by GEvo, a URL will be created that will be a hyperlink to GEvo with your analysis pre-configured. To regenerated the results, all you need to do is press the "Run GEvo Analysis!" button and wait for the analysis to run. This link is stored in two places:
- At the bottom of the results under "GEvo Links" (see example image.) This link has been condensed using the tinyurl redirecting service.
- At the bottom of the log file. The link to the log file can also be found at the bottom of the results (see example image.)
GEvo Direct
GEvo Direct is a tool for quickly viewing the results of a previously run analysis without having to re-run the analysis. Please Note: CoGe saves all the files from a GEvo analysis for ~24 hours. After that time, the data-files are deleted and the GEvo Direct link will no longer work.
Save Work History
Registered CoGe users can save a link to a GEvo analysis for later retrieval from their work history. This permits a GEvo analysis to also be names and annotated for future reference.
Modifying result graphics
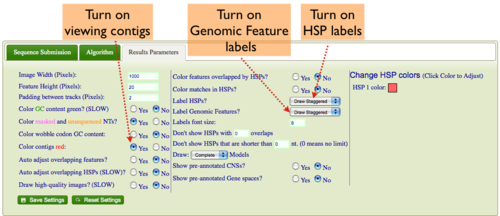
Showing Contigs
Some genomes have contig assembly information. To view this in GEvo's results:
- Select the "Results Parameters" tab from GEvo's configuration box
- Select "yes" for the option "Color contigs red".
Turning on labels for HSPs (blast hits) in GEvo's results
If you want to have the HSP number drawn on the HSP:
- Select the "Results Parameters" tab from GEvo's configuration box
- Select "yes" for the option "Label HSPs".
- You can have the labels drawn linearly, so each label is at the same vertical position for a track, or staggered, where they are drawn top, middle, bottom alternating.
Turning on labels for Genomic Features (e.g. genes) in GEvo's results
If you want to have the feature names drawn on the feature:
- Select the "Results Parameters" tab from GEvo's configuration box
- Select "yes" for the option "Label Genomic Features".
- You can have the labels drawn linearly, so each label is at the same vertical position for a track, or staggered, where they are drawn top, middle, bottom alternating.
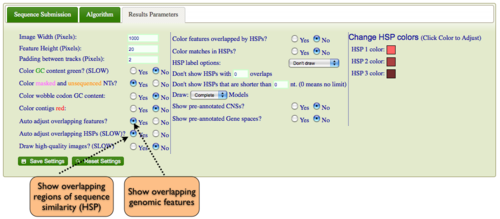
Expanding Overlapping Features and Regions of Sequence Similarity
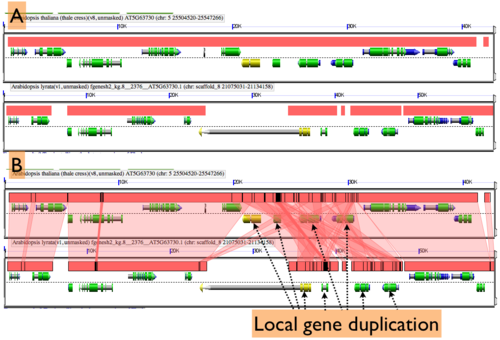
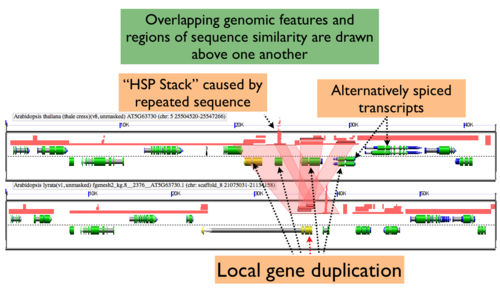
By default GEvo will drawn overlapping genomic features and regions of sequence similarity on top of one another. However, this sometimes hides some of the interesting complexities in a genomic region such as local duplications or regions containing repeated sequences. To view these, select the "Results Parameters" tab and select "Yes" for "Auto adjust overlapping features" and/or "Auto adjust overlapping HSPs". These options are set to "No" by default because finding and drawing overlapping features can take a long time to process, and are not always useful.
Merging Analyses
Often, there are times when you will want to merge together two or more separate GEvo anlayses. To do this, copy a GEvo link into the text-box next the text: "Merge Previous GEvo Analysis (paste in URL)" located at the top of the sequence submission tab. Then press the "Merge" button". The sequences as specified in the pasted URL will appear as new sequence submission boxes configured as specified in the link (extra up/downstream sequence, reverse complement, masked, etc.)
Refining an analysis
Once a GEvo analysis has run, you can change any of the analysis parameters and re-run the analysis by pressing the "Run GEvo analysis" button again. The common parameters changed are:
- The extent of the genomic region analyzed. The interactive results make this easy with slider bars.
- The algorithm used in the analysis
- Masking sequences
- Skipping sequences
- Reverse complementing sequences
- The coloration and information displayed in the result's graphics
Hints and Tricks
Sequences with many common sub-sequences
Comparing sequences with lots of common sub-sequences usually causes GEvo to take a very long time processing the analysis (both in terms of identifying the common sequences and generating the final results). Also, if many regions are identified, it is often difficult to make sense of the results. This kind of problem will surface in many large genomes, such as mammal and plant genomes. For example human and maize are both riddled with large amounts of repetitive sequences derived from retroviruses and transposons. This makes the comparison of large genome regions in these genomes difficult, if not impossible. To circumvent this problem, mask all sequence that does not code for protein. You can select this option under the "Sequence options" menu and selecting "non-CDS" for the row "Mask Sequence".
Example Analyses
Analysis of syntenic regions from Arabidopsis thaliana, Carica papaya, and Vitis vinifera
Linking to GEvo
Linking to GEvo is easy! Please see this page for how to do this.
Exporting Images for Publication
Please see exporting GEvo results for publication for how to do this.