Difference between revisions of "Utricularia and Genlisea pseudoassembly"
From CoGepedia
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | ==Overview== | ||
| + | [[Pseudoassemblies]] for both Utricularia and Genlisea are generated in [[SynMap]] using the [[Syntenic Path Assembly]] to the tomato genome. | ||
| + | |||
==High resolution analyses== | ==High resolution analyses== | ||
| Line 4: | Line 7: | ||
[[File:Screen shot 2012-02-06 at 9.55.40 AM.png|thumb|center|600px|Both Genlisea and Utricularia are pseudoassemblies against the tomato genome. Compared to syntenic tomato genomic regions. http://genomevolution.org/r/4h43]] | [[File:Screen shot 2012-02-06 at 9.55.40 AM.png|thumb|center|600px|Both Genlisea and Utricularia are pseudoassemblies against the tomato genome. Compared to syntenic tomato genomic regions. http://genomevolution.org/r/4h43]] | ||
| + | [[File:Screen shot 2012-02-06 at 3.07.02 PM.png|thumb|center|600px|Both Genlisea and Utricularia are pseudoassemblies against the tomato genome. Compared to syntenic tomato genomic regions. '''Note: All HSPs drawn on top''' http://genomevolution.org/r/4h6p]] | ||
==Sytnenic dotplots== | ==Sytnenic dotplots== | ||
[[File:Screen Shot 2012-02-06 at 6.26.10 AM.png|thumb|left|600px|SynMap analysis of pseudoassembly of Utricularia's contigs (based on synteny to tomato; y-axis) to tomato (x-axis). Results may be regenerated at http://genomevolution.org/r/4h1z]] | [[File:Screen Shot 2012-02-06 at 6.26.10 AM.png|thumb|left|600px|SynMap analysis of pseudoassembly of Utricularia's contigs (based on synteny to tomato; y-axis) to tomato (x-axis). Results may be regenerated at http://genomevolution.org/r/4h1z]] | ||
Latest revision as of 09:47, 13 February 2012
Overview
Pseudoassemblies for both Utricularia and Genlisea are generated in SynMap using the Syntenic Path Assembly to the tomato genome.
High resolution analyses
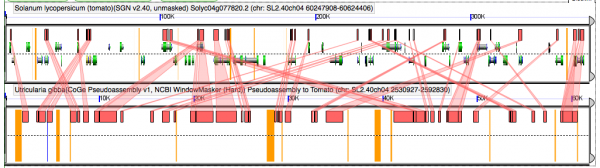
GEvo analysis of pseudoassembly Utricularia to tomato. The pseudoassembly of Utricularia is based on the syntenic path assembly to tomato. As such, duplicated utricularia genomic regions will appear next to each other. If Utricularia has had an independent WGD followed by fractionation, these neighboring pieces will show intercalated synteny to tomato. Results may be regenerated at http://genomevolution.org/r/4h2c

Both Genlisea and Utricularia are pseudoassemblies against the tomato genome. Compared to syntenic tomato genomic regions. http://genomevolution.org/r/4h43

Both Genlisea and Utricularia are pseudoassemblies against the tomato genome. Compared to syntenic tomato genomic regions. Note: All HSPs drawn on top http://genomevolution.org/r/4h6p
Sytnenic dotplots

SynMap analysis of pseudoassembly of Utricularia's contigs (based on synteny to tomato; y-axis) to tomato (x-axis). Results may be regenerated at http://genomevolution.org/r/4h1z