Difference between revisions of "LoadAnnotation"
From CoGepedia
| Line 13: | Line 13: | ||
===Data File=== | ===Data File=== | ||
| − | Annotations are specified in GFF format | + | Annotations are specified in GFF format. It is VERY IMPORTANT that the GFF format follows CoGe guidelines, please see see [[GFF ingestion]] for details. |
| + | |||
| + | You can select and retrieve the GFF file from multiple sources: | ||
*The iPlant Data Store | *The iPlant Data Store | ||
*A URL (FTP/HTTP) server | *A URL (FTP/HTTP) server | ||
*Your computer (Upload) | *Your computer (Upload) | ||
Revision as of 12:45, 17 April 2013
LoadAnnotation enables you to load a set of annotation data for a genome in CoGe. The genome and its annotations can then be viewed graphically in GenomeView.
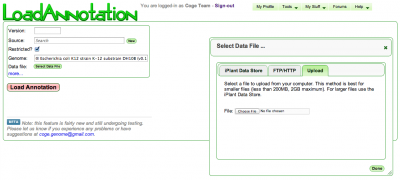
Input
- Version: Version of annotation
- Source: Where is the data from? This could be you, your lab, your university, a sequencing center, your collaborator.
- Restricted: Is this annotation public or restricted to you and your collaborators
- Genome: Select the appropriate genome from CoGe
- Select Data File: Opens a window for specifying the input data file
- Name: Name of annotation data set (optional)
- Description: Description of annotation data set (optional)
Data File
Annotations are specified in GFF format. It is VERY IMPORTANT that the GFF format follows CoGe guidelines, please see see GFF ingestion for details.
You can select and retrieve the GFF file from multiple sources:
- The iPlant Data Store
- A URL (FTP/HTTP) server
- Your computer (Upload)