Difference between revisions of "GEvo-4at-cp-vv"
| Line 4: | Line 4: | ||
=Comparison of syntenic regions among ''Arabidopsis thaliana'', ''Carica papaya'', and ''Vitis vinifera''= | =Comparison of syntenic regions among ''Arabidopsis thaliana'', ''Carica papaya'', and ''Vitis vinifera''= | ||
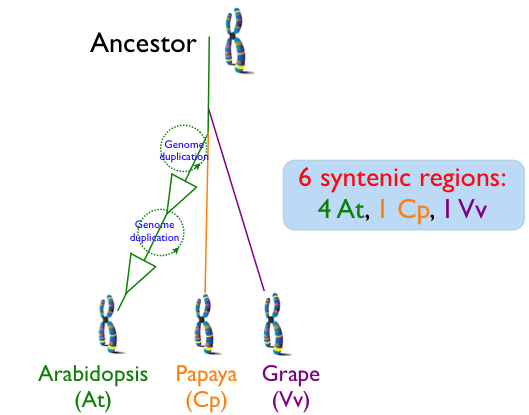
| − | Since the divergence of ''Arabidopsis thaliana'' (At), ''Carica papaya'' (Cp; papaya), and ''Vitis vinifera'' (Vv; grape), the Arabidopsis lineage has undergone two sequential whole genome duplication events, while the genomes of papaya and grape have not. This means that for every genomic regions of grape, the most basal of these lineages, there is one corresponding syntenic region in papaya, and 4 corresponding syntenic regions in arabidopsis. While whole genome duplication events contemporaneously create a copy of every chromosome and all their underlying [[genomic features]] (e.g. genes), over evolutionary time, many of the duplicated features are lost from [[homeologous]] region or its partner region. While this diploidization process will tend to return a genome's gene content to one that is more similar to the pre-[[polyploid]] ancestor, the overall structure of the genome will be changed as formerly neighboring genes may be dispersed on different derived homeologous chromosomes. This process of homeologous gene loss following whole genome duplication events is known as [[fractionation]]. Comparison of fractionated homeologs to an outgroup syntenic genomic region that has not had its own separate whole genome duplication event will yield an expected pattern: the outgroup genomic region will contain nearly the entire gene content of the fractionated syntenic regions, and the genomic arrangement of these genes will be collinear. In addition, the outgroup syntelogs of the fractionated homeologs will be intercalated with respect to one another. | + | Since the divergence of ''Arabidopsis thaliana'' (At), ''Carica papaya'' (Cp; papaya), and ''Vitis vinifera'' (Vv; grape), the Arabidopsis lineage has undergone two sequential whole genome duplication events, while the genomes of papaya and grape have not. This means that for every genomic regions of grape, the most basal of these lineages, there is one corresponding syntenic region in papaya, and 4 corresponding syntenic regions in arabidopsis. While whole genome duplication events contemporaneously create a copy of every chromosome and all their underlying [[genomic features]] (e.g. genes), over evolutionary time, many of the duplicated features are lost from [[homeologous]] region or its partner region. While this diploidization process will tend to return a genome's gene content to one that is more similar to the pre-[[polyploid]] ancestor, the overall structure of the genome will be changed as formerly neighboring genes may be dispersed on different derived homeologous chromosomes. This process of homeologous gene loss following whole genome duplication events is known as [[fractionation]]. Comparison of fractionated homeologs to an outgroup syntenic genomic region that has not had its own separate whole genome duplication event will yield an expected pattern: the outgroup genomic region will contain nearly the entire gene content of the fractionated syntenic regions, and the genomic arrangement of these genes will be collinear. In addition, the outgroup syntelogs of the fractionated homeologs will be intercalated with respect to one another. While fractionation may be a dominate mechanism in post-polyploid genome evolution, other mechanisms, such as gene [[transposition]] and local duplication events, are also at play that can decrease the syntenic signal by moving DNA into and out of a genomic region. |
| + | |||
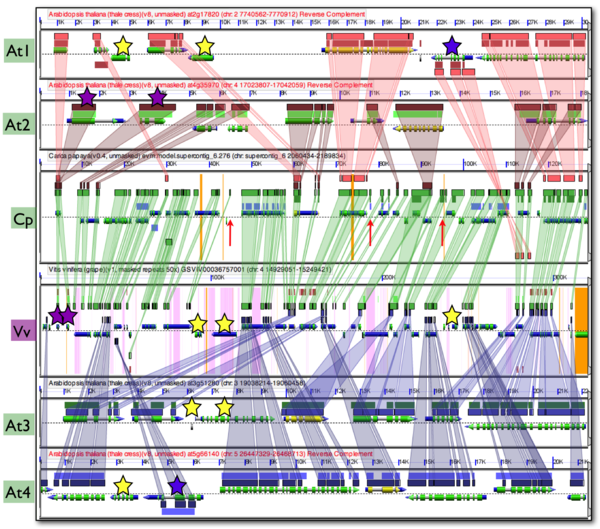
| + | The figure shown here is a syntenic comparison of 4 Arabidopsis regions (At1, At2, At3, Agt4) to an outgroup region of papaya (Cp), and a second outgroup region of grape (Vv). Each genomic region is represented by a panel visualizing the genomic region. The dashed line in the middle of a panel separates the top (5' left) and bottom (3' left) strand of DNA. Gene models are drawn above and below this line for each strand respectively. Gene models are colored composite arrows where the narrow gray arrow represents the extent of the gene model, blue represents mRNA, and green represents coding sequence (CDS). Other genomic features, such as tRNAs | ||
Revision as of 18:12, 25 August 2009
Comparison of syntenic regions among Arabidopsis thaliana, Carica papaya, and Vitis vinifera
Since the divergence of Arabidopsis thaliana (At), Carica papaya (Cp; papaya), and Vitis vinifera (Vv; grape), the Arabidopsis lineage has undergone two sequential whole genome duplication events, while the genomes of papaya and grape have not. This means that for every genomic regions of grape, the most basal of these lineages, there is one corresponding syntenic region in papaya, and 4 corresponding syntenic regions in arabidopsis. While whole genome duplication events contemporaneously create a copy of every chromosome and all their underlying genomic features (e.g. genes), over evolutionary time, many of the duplicated features are lost from homeologous region or its partner region. While this diploidization process will tend to return a genome's gene content to one that is more similar to the pre-polyploid ancestor, the overall structure of the genome will be changed as formerly neighboring genes may be dispersed on different derived homeologous chromosomes. This process of homeologous gene loss following whole genome duplication events is known as fractionation. Comparison of fractionated homeologs to an outgroup syntenic genomic region that has not had its own separate whole genome duplication event will yield an expected pattern: the outgroup genomic region will contain nearly the entire gene content of the fractionated syntenic regions, and the genomic arrangement of these genes will be collinear. In addition, the outgroup syntelogs of the fractionated homeologs will be intercalated with respect to one another. While fractionation may be a dominate mechanism in post-polyploid genome evolution, other mechanisms, such as gene transposition and local duplication events, are also at play that can decrease the syntenic signal by moving DNA into and out of a genomic region.
The figure shown here is a syntenic comparison of 4 Arabidopsis regions (At1, At2, At3, Agt4) to an outgroup region of papaya (Cp), and a second outgroup region of grape (Vv). Each genomic region is represented by a panel visualizing the genomic region. The dashed line in the middle of a panel separates the top (5' left) and bottom (3' left) strand of DNA. Gene models are drawn above and below this line for each strand respectively. Gene models are colored composite arrows where the narrow gray arrow represents the extent of the gene model, blue represents mRNA, and green represents coding sequence (CDS). Other genomic features, such as tRNAs